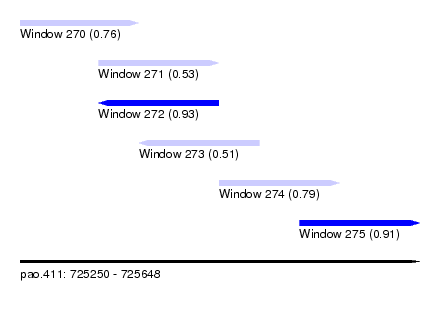
| Sequence ID | pao.411 |
|---|---|
| Location | 725,250 – 725,648 |
| Length | 398 |
| Max. P | 0.930555 |

| Location | 725,250 – 725,368 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.80 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -31.67 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725250 118 + 6264404 CUGCGGGUGUCACGUAAG--UGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGG ((((..(((((((....)--)))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......))))........... ( -38.00) >pau.2632 1583625 118 - 6537648 CUGCGGGUGUCACGUAAG--UGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGG ((((..(((((((....)--)))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......))))........... ( -38.00) >pss.496 967921 120 + 6093698 CUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGG ((((..((((((((....)))))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......))))........... ( -35.40) >pfo.1282 2552723 118 + 6438405 CUACGGGUAUCACGCAAG--UGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGG ((((..(((((((....)--)))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......))))........... ( -37.50) >consensus CUACGGGUAUCACGUAAG__UGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGG ((((..((((((........)))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......))))........... (-31.67 = -30.93 + -0.75)

| Location | 725,328 – 725,448 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -37.95 |
| Energy contribution | -37.70 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725328 120 + 6264404 UAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGA ..(((..(((......)))....)))..........(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....)))). ( -36.80) >pau.2632 1583703 120 - 6537648 UAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGA ..(((..(((......)))....)))..........(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....)))). ( -36.80) >pss.496 968001 120 + 6093698 UAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGA ..(((..(((......)))....)))..........(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....)))). ( -38.60) >pfo.1282 2552801 120 + 6438405 UAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGA ..(((..(((......)))....)))..........(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....)))). ( -38.60) >consensus UAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGA ..(((..(((......)))....)))..........(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....)))). (-37.95 = -37.70 + -0.25)


| Location | 725,328 – 725,448 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -39.30 |
| Energy contribution | -39.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725328 120 - 6264404 UCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUA .((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).............((((((.(........))))))) ( -39.30) >pau.2632 1583703 120 + 6537648 UCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUA .((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).............((((((.(........))))))) ( -39.30) >pss.496 968001 120 - 6093698 UCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUA .((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).............((((((.(........))))))) ( -41.80) >pfo.1282 2552801 120 - 6438405 UCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUA .((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).............((((((.(........))))))) ( -41.80) >consensus UCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUA .((((..(((.(((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).............((((((.(........))))))) (-39.30 = -39.30 + 0.00)

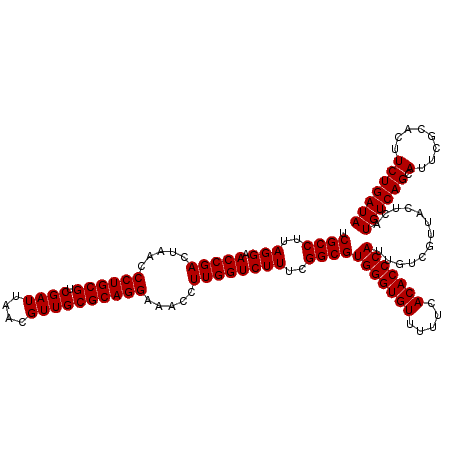
| Location | 725,368 – 725,488 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -34.55 |
| Energy contribution | -35.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725368 120 - 6264404 GGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACA ((((...........))))................((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).))......... ( -35.00) >pau.2632 1583743 120 + 6537648 GGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACA ((((...........))))................((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).))......... ( -35.00) >pss.496 968041 120 - 6093698 GGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACA ((((.((......)).))))...............((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).))......... ( -39.20) >pfo.1282 2552841 120 - 6438405 GGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACA ((((.((......)).))))...............((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).))......... ( -39.20) >consensus GGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACA ((((.((......)).))))...............((.((.((((..(((.(((((.....((((((.((((....)))))))))).....))))))))..)))).)).))......... (-34.55 = -35.05 + 0.50)


| Location | 725,448 – 725,568 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -36.86 |
| Energy contribution | -36.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.787076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725448 120 + 6264404 GGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGG .(((....)))(((((.(((.(((((((((........)))))..)))).)))))))).......((((((..((.((((((....)))))).))..))))))................. ( -40.30) >pau.2632 1583823 120 - 6537648 GGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGG .(((....)))(((((.(((.(((((((((........)))))..)))).)))))))).......((((((..((.((((((....)))))).))..))))))................. ( -40.30) >pss.496 968121 120 + 6093698 GGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGG .(((....))).........((((.(((((........)))))..))))..((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))). ( -38.10) >pfo.1282 2552921 120 + 6438405 GGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGG .(((....))).........((((.(((((........)))))..))))..((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))). ( -35.40) >consensus GGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGG .(((...(((((((((.(((.(((((((((........)))))..)))).)))))))).......((((((..((.((((((....)))))).))..))))))..))))..)))...... (-36.86 = -36.93 + 0.06)

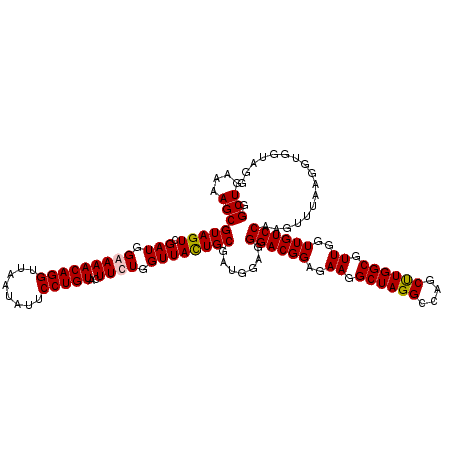
| Location | 725,528 – 725,648 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -37.66 |
| Energy contribution | -37.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725528 120 + 6264404 GGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGC .((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))....((.(((.((..((((((.....))))))...)).)))...)) ( -39.70) >pau.2632 1583903 120 - 6537648 GGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGC .((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))....((.(((.((..((((((.....))))))...)).)))...)) ( -39.70) >pss.496 968201 120 + 6093698 GGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACACGAAGUGGUUGAUGC (((((((((((((.....)).))))))).............(((((((((.((.....))))))))))).))))....((.(((.((..((((((.....))))))...)).)))...)) ( -41.40) >pfo.1282 2553001 119 + 6438405 GGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGU-UGCCUUUAGGCGACGAAGUGGUUGAUGC .((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))....((.(((.((..((-((((....))))))...)).)))...)) ( -41.50) >consensus GGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGC .((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))....((.(((.((..((((((.....))))))...)).)))...)) (-37.66 = -37.23 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:13 2007