
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,039,536 – 6,039,654 |
| Length | 118 |
| Max. P | 0.997970 |

| Location | 6,039,536 – 6,039,654 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -43.64 |
| Consensus MFE | -38.81 |
| Energy contribution | -39.03 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
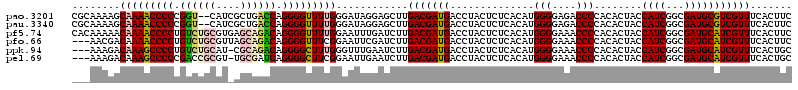
>pao.3201 6039536 118 + 6264404 CGCAAAAGCAAAACCCCCGGU--CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUC ........(((((((((.(((--((....))))).))))))))).....((((...(((((((..............(((....)))........((((...)))))))))))...)))) ( -41.40) >pau.3340 6312455 118 + 6537648 CGCAAAAGCAAAACCCCCGGU--CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUC ........(((((((((.(((--((....))))).))))))))).....((((...(((((((..............(((....)))........((((...)))))))))))...)))) ( -41.40) >pf5.74 6946598 120 - 7074893 CACAAAAACAAAACCCCUGUCUGCGUGAGCAGACAGGGGUUUUGGAAUUUGAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUC ..((((..((((((((((((((((....))))))))))))))))...)))).....(((((((..............(((....)))........((((...)))))))))))....... ( -52.30) >pfo.66 6311451 117 - 6438405 ---AACGACAAAACCCCUGUCUGCGUUAGCAGACAGGGGUUUCGGAAUUCGAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUC ---..(((...(((((((((((((....))))))))))))))))(((..((((((((.(((((..............(((....)))........))))).))).).)))).)))..... ( -44.55) >ppk.94 5999808 116 - 6181863 ---AAAGACAAAGCCCCUGUCUGCAU-CGCAGACAGGGGCUUUGGGUUUGAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUGC ---(((..((((((((((((((((..-.))))))))))))))))..))).......(((((((..............(((....)))........((((...)))))))))))....... ( -48.40) >pel.69 5767854 116 - 5888780 ---AAAGACAAAGCCCCCGACCGCGU-UGCGAUCAGGGGCUUCGGAAUUGAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUGC ---.....(.(((((((.((.(((..-.))).)).))))))).)............(((((((..............(((....)))........((((...)))))))))))....... ( -33.80) >consensus ___AAAAACAAAACCCCCGUCUGCAU_AGCAAACAGGGGUUUUGGAAUUGGAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUC ........(((((((((.((((((....)))))).)))))))))............(((((((..............(((....)))........((((...)))))))))))....... (-38.81 = -39.03 + 0.22)
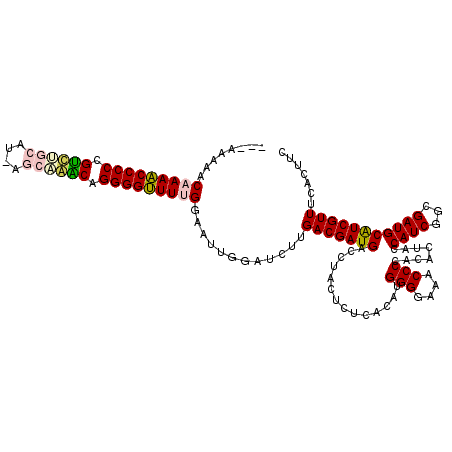
| Location | 6,039,536 – 6,039,654 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -50.20 |
| Consensus MFE | -47.88 |
| Energy contribution | -46.50 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
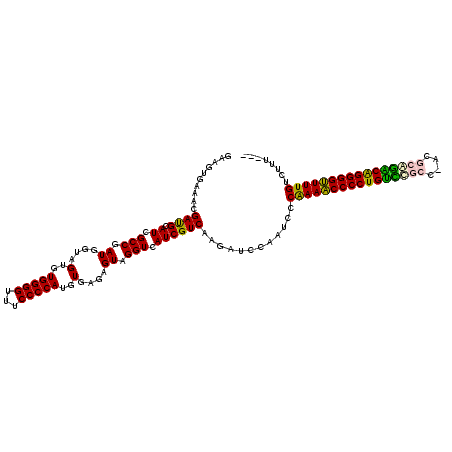
>pao.3201 6039536 118 - 6264404 GAAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG--ACCGGGGGUUUUGCUUUUGCG ...(..(((.((((.((.(((.((....(..(((((...)))))..)....)).))).))))))............(((((((((((((((....))--))))))))))))).)))..). ( -49.70) >pau.3340 6312455 118 - 6537648 GAAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG--ACCGGGGGUUUUGCUUUUGCG ...(..(((.((((.((.(((.((....(..(((((...)))))..)....)).))).))))))............(((((((((((((((....))--))))))))))))).)))..). ( -49.70) >pf5.74 6946598 120 + 7074893 GAAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCAAAUUCCAAAACCCCUGUCUGCUCACGCAGACAGGGGUUUUGUUUUUGUG ........((((((....(((.((....(..(((((...)))))..)....)).)))))))))......((((...((((((((((((((((....))))))))))))))))..)))).. ( -50.40) >pfo.66 6311451 117 + 6438405 GAAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCGAAUUCCGAAACCCCUGUCUGCUAACGCAGACAGGGGUUUUGUCGUU--- ((..(((..(((((....(((.((....(..(((((...)))))..)....)).)))))))))))...)).(((..((((((((((((((((....))))))))))))))))..)))--- ( -51.70) >ppk.94 5999808 116 + 6181863 GCAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUCAAACCCAAAGCCCCUGUCUGCG-AUGCAGACAGGGGCUUUGUCUUU--- ....((((((((((....(((.((....(..(((((...)))))..)....)).))))))))).....))))....((((((((((((((((.-..)))))))))))))))).....--- ( -50.90) >pel.69 5767854 116 + 5888780 GCAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUCAAUUCCGAAGCCCCUGAUCGCA-ACGCGGUCGGGGGCUUUGUCUUU--- ....((((((((((....(((.((....(..(((((...)))))..)....)).))))))))).....))))....((((((((((((((((.-..)))))))))))))))).....--- ( -48.80) >consensus GAAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCCAAUCCCAAAACCCCUGUCCGCC_ACGCAGACAGGGGUUUUGUCUUU___ ..........((((.((.(((.((....(..(((((...)))))..)....)).))).))))))............((((((((((((((((....))))))))))))))))........ (-47.88 = -46.50 + -1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:27 2007