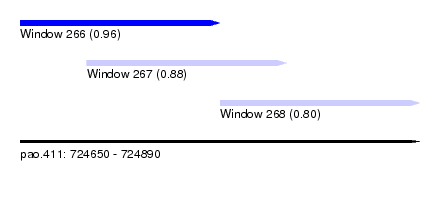
| Sequence ID | pao.411 |
|---|---|
| Location | 724,650 – 724,890 |
| Length | 240 |
| Max. P | 0.955785 |

| Location | 724,650 – 724,770 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -40.31 |
| Energy contribution | -39.81 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
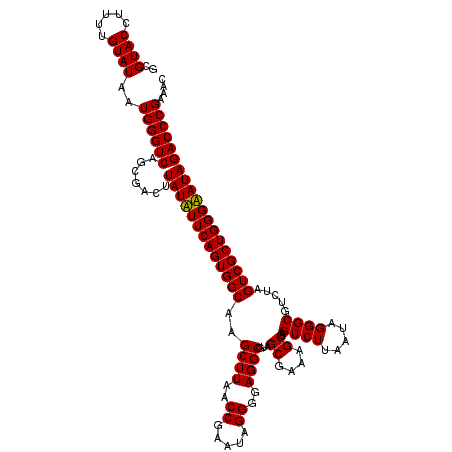
>pao.411 724650 120 + 6264404 GCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUUUAGUCGCUGGGUAUAGACCCGAAAC ..((((.....))))..((((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).....))))))))))))))))))).... ( -40.81) >pau.2632 1583025 120 - 6537648 GCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUUUAGUCGCUGGGUAUAGACCCGAAAC ..((((.....))))..((((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).....))))))))))))))))))).... ( -40.81) >pss.496 967321 120 + 6093698 GCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAAC ..((((.....))))..((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).))..)))))))..)))))))))).... ( -41.51) >pfo.1282 2552123 120 + 6438405 GCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAAC ..((((.....))))..((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).))..)))))))..)))))))))).... ( -41.51) >consensus GCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAAC ..((((.....))))..((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))))))).... (-40.31 = -39.81 + -0.50)

| Location | 724,690 – 724,810 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -39.54 |
| Energy contribution | -39.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724690 120 + 6264404 GGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACA .....((((((((.....((.(((((...((....))..((((.....))))))))).))(((.((.(((((((((....))))...)))))))...)))........)))))))).... ( -39.90) >pau.2632 1583065 120 - 6537648 GGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACA .....((((((((.....((.(((((...((....))..((((.....))))))))).))(((.((.(((((((((....))))...)))))))...)))........)))))))).... ( -39.90) >pss.496 967361 120 + 6093698 GGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACA (((((((((..((.....))..))))...((....))((((((.....)))).))...)))))........(((((..(((...((((((..(....)..))))))..)))))))).... ( -41.00) >pfo.1282 2552163 120 + 6438405 GGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACA .....((((((((.....((.(((((...((....))..))))).....((.(((((.((.....)).)))))))....))...((((((..(....)..))))))..)))))))).... ( -40.10) >consensus GGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGUCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACA .....((((((((.....((.(((((...((....))..((((.....))))))))).))(((.((.(((((((((....))))...)))))))...)))........)))))))).... (-39.54 = -39.35 + -0.19)

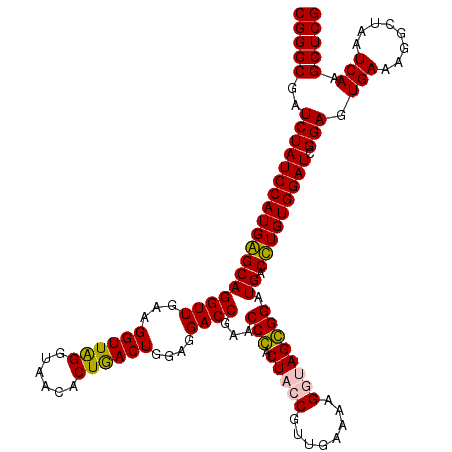
| Location | 724,770 – 724,890 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -40.78 |
| Energy contribution | -40.40 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
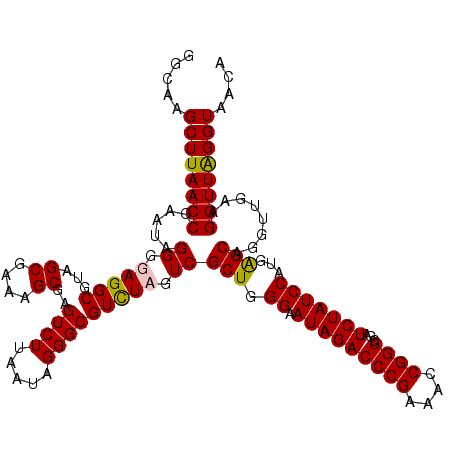
>pao.411 724770 120 + 6264404 CGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCG (((((..((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).))).(((........))).))))) ( -39.90) >pau.2632 1583145 120 - 6537648 CGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCG (((((..((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).))).(((........))).))))) ( -39.90) >pss.496 967441 120 + 6093698 CGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCG (((((..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).(((........))).))))) ( -40.20) >pfo.1282 2552243 120 + 6438405 CGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCG (((((..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).(((........))).))))) ( -40.60) >consensus CGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUACCGUUGAAAAGGUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCG (((((..((((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))))))))).))).(((........))).))))) (-40.78 = -40.40 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:05 2007