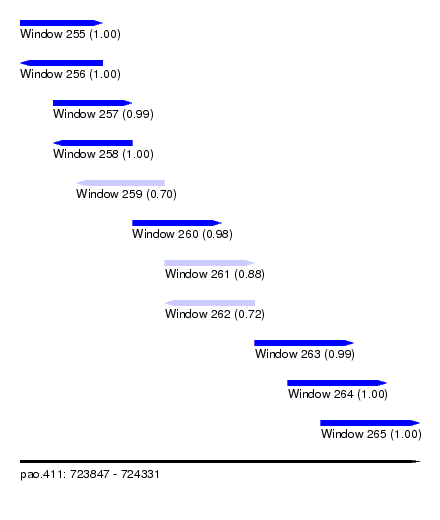
| Sequence ID | pao.411 |
|---|---|
| Location | 723,847 – 724,331 |
| Length | 484 |
| Max. P | 0.999572 |

| Location | 723,847 – 723,947 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -26.15 |
| Energy contribution | -23.21 |
| Covariance contribution | -2.94 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723847 100 + 6264404 CAGGAGUUCGAUCCUCCUUGGCUCCACCAUCUAAAACAAUCG--UCGAAAGCUCAGAAAUGAAUGUUC----------GUGAA--------UGAACAUUGAUUUCUGGUCUUUGCACCAG ..(((((.(((......))))))))................(--(((((..(.(((((((.(((((((----------(....--------)))))))).))))))))..)))).))... ( -29.30) >pau.2632 1582222 100 - 6537648 CAGGAGUUCGAUCCUCCUUGGCUCCACCAUCUAAAACAAUCG--UCGAAAGCUCAGAAAUGAAUGUUC----------GUGAA--------UGAACAUUGAUUUCUGGUCUUUGCACCAG ..(((((.(((......))))))))................(--(((((..(.(((((((.(((((((----------(....--------)))))))).))))))))..)))).))... ( -29.30) >pss.496 966500 119 + 6093698 CAGCGGUUCGAUCCCGCUUGGCUCCACCACUUACUGCUUCUGUUUUGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCCUGAAUGUUGAUUUCUAGUCUUUG-AUUAG .(((((.......)))))(((.....)))..........(((...(.((((.((((((((.(((((((((((((((.....))))))))...))))))).)))))))).)))).-).))) ( -34.40) >pfo.1282 2551301 109 + 6438405 CAGCGGUUCGAUCCCGCUUGGCUCCACCAC-UACUGCUUCUGAAGUAAGAGCUUAGAAAUGAGCAUUCCAUCGCUGC-GAUGG--------UGAAUGUUGAUUUCUAGUCUUUG-ACUAG .(((((.......)))))(((.....))).-............(((.((((.((((((((.((((((((((((...)-)))))--------..)))))).)))))))).)))).-))).. ( -32.50) >consensus CAGCAGUUCGAUCCCCCUUGGCUCCACCACCUAAAACAACCG__UCGAAAGCUCAGAAAUGAACAUUC__________GAGAA________UGAACAUUGAUUUCUAGUCUUUG_ACCAG .(((((.......)))))(((.....)))..................((((.((((((((.(((((((........................))))))).)))))))).))))....... (-26.15 = -23.21 + -2.94)

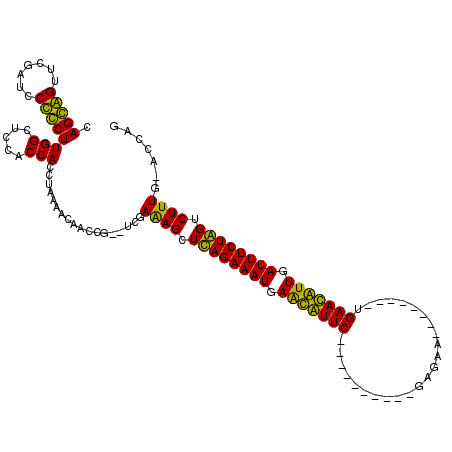
| Location | 723,847 – 723,947 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -24.16 |
| Energy contribution | -22.41 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723847 100 - 6264404 CUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUCAC----------GAACAUUCAUUUCUGAGCUUUCGA--CGAUUGUUUUAGAUGGUGGAGCCAAGGAGGAUCGAACUCCUG .((((...(((..(((((((.(((((((.--------.....----------))))))).)))))))..)))((.(--(.(((......))).)).))))))(((((.......))))). ( -32.60) >pau.2632 1582222 100 + 6537648 CUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUCAC----------GAACAUUCAUUUCUGAGCUUUCGA--CGAUUGUUUUAGAUGGUGGAGCCAAGGAGGAUCGAACUCCUG .((((...(((..(((((((.(((((((.--------.....----------))))))).)))))))..)))((.(--(.(((......))).)).))))))(((((.......))))). ( -32.60) >pss.496 966500 119 - 6093698 CUAAU-CAAAGACUAGAAAUCAACAUUCAGGCACUGAUCAUCAGGAUCAAUGGAAUGCUCAUUUCUAAGCUUUCAAAACAGAAGCAGUAAGUGGUGGAGCCAAGCGGGAUCGAACCGCUG ((.((-((...(((((((((...(((((...((.(((((.....))))).)))))))...))))))).(((((.......))))).))...))))..))...(((((.......))))). ( -29.40) >pfo.1282 2551301 109 - 6438405 CUAGU-CAAAGACUAGAAAUCAACAUUCA--------CCAUC-GCAGCGAUGGAAUGCUCAUUUCUAAGCUCUUACUUCAGAAGCAGUA-GUGGUGGAGCCAAGCGGGAUCGAACCGCUG (((((-(...)))))).........((((--------((((.-.(((.(((((.....))))).))..(((((......)).))).)..-))))))))....(((((.......))))). ( -32.20) >consensus CUAGU_CAAAGACCAGAAAUCAACAUUCA________UCAAC__________GAACACUCAUUUCUAAGCUUUCAA__CAAAAGCAGUAGAUGGUGGAGCCAAGCAGGAUCGAACCCCUG .......((((..(((((((.(((((((........................))))))).)))))))..))))..................(((.....)))(((((.......))))). (-24.16 = -22.41 + -1.75)

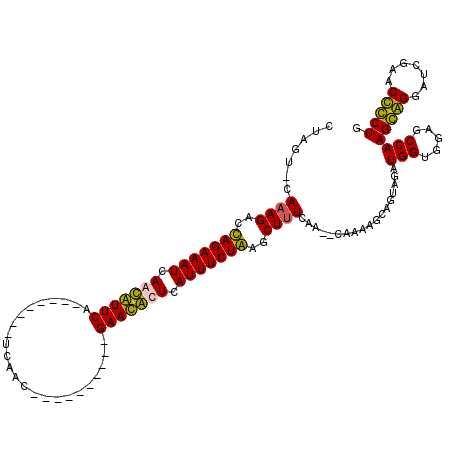
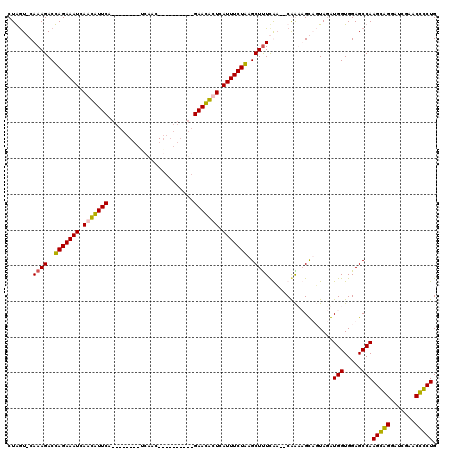
| Location | 723,887 – 723,983 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.75 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -18.45 |
| Energy contribution | -16.51 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723887 96 + 6264404 CG--UCGAAAGCUCAGAAAUGAAUGUUC----------GUGAA--------UGAACAUUGAUUUCUGGUCUUUGCACCAGAACUGUUCUUUAAAAAUUCGGGUAUGUGAUAGAAGU---- .(--(((((((..(((((((.(((((((----------(....--------)))))))).)))))))..))))..(((.(((..............))).)))...))))......---- ( -24.34) >pau.2632 1582262 96 - 6537648 CG--UCGAAAGCUCAGAAAUGAAUGUUC----------GUGAA--------UGAACAUUGAUUUCUGGUCUUUGCACCAGAACUGUUCUUUAAAAAUUCGGGUAUGUGAUAGAAGU---- .(--(((((((..(((((((.(((((((----------(....--------)))))))).)))))))..))))..(((.(((..............))).)))...))))......---- ( -24.34) >pss.496 966540 118 + 6093698 UGUUUUGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCCUGAAUGUUGAUUUCUAGUCUUUG-AUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUAGAAAGAAAU .....(.((((.((((((((.(((((((((((((((.....))))))))...))))))).)))))))).)))).-).......-.((((((....((........)).....)))))).. ( -29.10) >pfo.1282 2551340 105 + 6438405 UGAAGUAAGAGCUUAGAAAUGAGCAUUCCAUCGCUGC-GAUGG--------UGAAUGUUGAUUUCUAGUCUUUG-ACUAGUUC-GUUCUUUAAAAAUUUGGGUAUGUGAUAGAAAG---- (((((...((((((((((((.((((((((((((...)-)))))--------..)))))).)))))))(((...)-)).)))))-...)))))........................---- ( -27.60) >consensus CG__UCGAAAGCUCAGAAAUGAACAUUC__________GAGAA________UGAACAUUGAUUUCUAGUCUUUG_ACCAGAAC_GUUCUUUAAAAAUUCGGGUAUGUGAUAGAAAG____ .......((((.((((((((.(((((((........................))))))).)))))))).))))............................................... (-18.45 = -16.51 + -1.94)

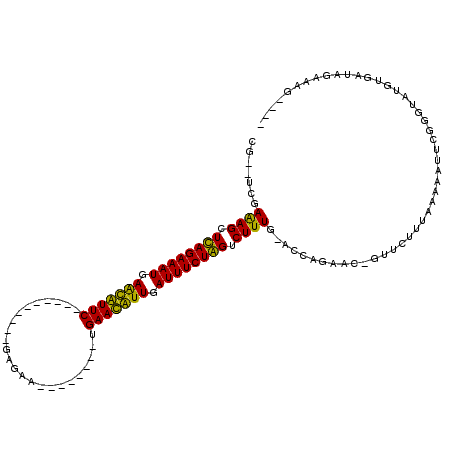
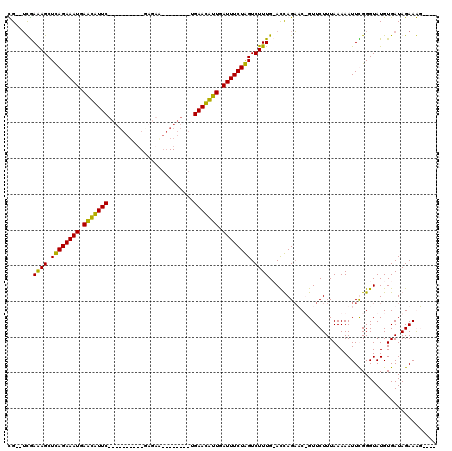
| Location | 723,887 – 723,983 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.75 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -13.06 |
| Energy contribution | -12.31 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723887 96 - 6264404 ----ACUUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUCAC----------GAACAUUCAUUUCUGAGCUUUCGA--CG ----.......((...((((.(((((.((....)).))))).)))).((((..(((((((.(((((((.--------.....----------))))))).)))))))..)))).))--.. ( -23.50) >pau.2632 1582262 96 + 6537648 ----ACUUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUCAC----------GAACAUUCAUUUCUGAGCUUUCGA--CG ----.......((...((((.(((((.((....)).))))).)))).((((..(((((((.(((((((.--------.....----------))))))).)))))))..)))).))--.. ( -23.50) >pss.496 966540 118 - 6093698 AUUUCUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAU-CAAAGACUAGAAAUCAACAUUCAGGCACUGAUCAUCAGGAUCAAUGGAAUGCUCAUUUCUAAGCUUUCAAAACA ...((.((((.....................)))).-))......-.((((..(((((((...(((((...((.(((((.....))))).)))))))...)))))))..))))....... ( -16.90) >pfo.1282 2551340 105 - 6438405 ----CUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAACUAGU-CAAAGACUAGAAAUCAACAUUCA--------CCAUC-GCAGCGAUGGAAUGCUCAUUUCUAAGCUCUUACUUCA ----..........................((((..-...(((((-(...)))))).............--------(((((-(...))))))...(((........)))))))...... ( -15.80) >consensus ____ACUUCUAUCACAUACCCAAAUUUUUAAAGAAC_GAUCUAGU_CAAAGACCAGAAAUCAACAUUCA________UCAAC__________GAACACUCAUUUCUAAGCUUUCAA__CA ...............................................((((..(((((((.(((((((........................))))))).)))))))..))))....... (-13.06 = -12.31 + -0.75)

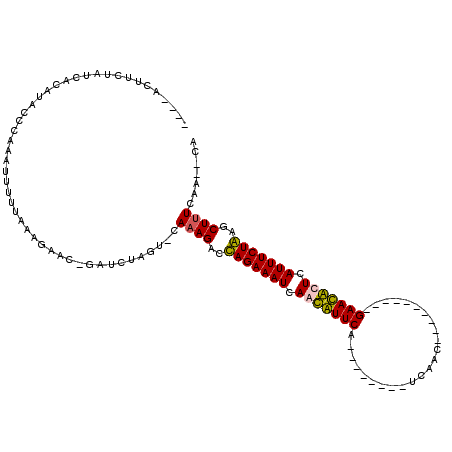
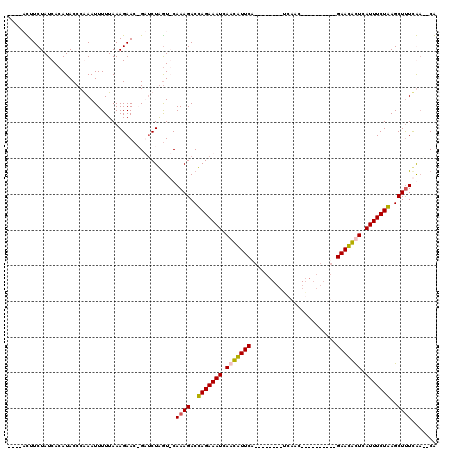
| Location | 723,915 – 724,022 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.32 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -10.73 |
| Energy contribution | -12.47 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723915 107 - 6264404 GACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCU-U----ACUUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUC ((((.((((((((.(........).)))))))))))).-.----............................(((((.(((((((......))))))).....))))).--------... ( -28.20) >pau.2632 1582290 107 + 6537648 GACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCU-U----ACUUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA--------UUC ((((.((((((((.(........).)))))))))))).-.----............................(((((.(((((((......))))))).....))))).--------... ( -28.20) >pss.496 966580 117 - 6093698 AGCCCGGACACACACCAGUGAAAGAGGUGCC-CGGUCUAUAUUUCUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAU-CAAAGACUAGAAAUCAACAUUCAGGCACUGAUCA .(((.((((((......))).....(((..(-..((((....((((((.....................)))))).-(((...))-)..))))..)..)))....))).)))........ ( -16.20) >pfo.1282 2551379 104 - 6438405 AGCCUGAUCAC-CACCAGUGAAAGUGGCCAU-CAGUCUAU----CUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAACUAGU-CAAAGACUAGAAAUCAACAUUCA--------CCA ((.(((((..(-(((........))))..))-))).)).(----(.((((.....................)))).-)).(((((-(...)))))).............--------... ( -17.20) >consensus AACCUGAACAAUCACCAGUGAAAGAGAUCAU_CAGUCU_U____ACUUCUAUCACAUACCCAAAUUUUUAAAGAAC_GAUCUAGU_CAAAGACCAGAAAUCAACAUUCA________UCA .................((((.((((....................)))).)))).................(((((.(((((((......))))))).....)))))............ (-10.73 = -12.47 + 1.75)

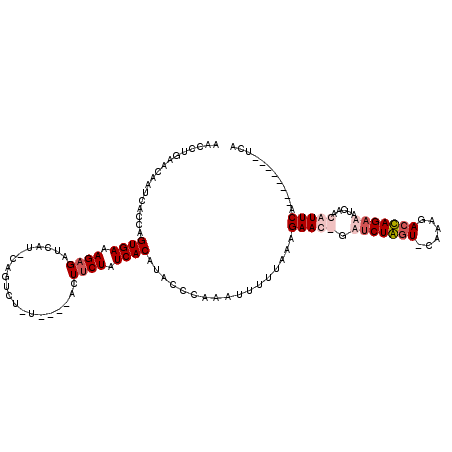
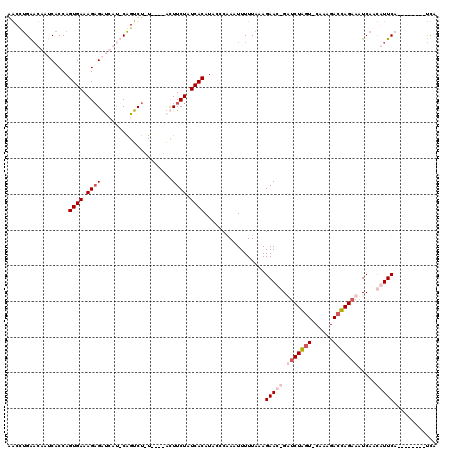
| Location | 723,983 – 724,091 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -19.07 |
| Energy contribution | -20.51 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723983 108 + 6264404 A-AGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUU-----------CAAGCGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCU .-.((((((((((((.((......)).)))))))).))))..(((((((((((((....-----------....)))))))))).((((.....))))...........)))........ ( -33.50) >pau.2632 1582358 108 - 6537648 A-AGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUU-----------CAAGCGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCU .-.((((((((((((.((......)).)))))))).))))..(((((((((((((....-----------....)))))))))).((((.....))))...........)))........ ( -33.50) >pss.496 966658 103 + 6093698 AUAGACCG-GGCACCUCUUUCACUGGUGUGUGUCCGGGCUAAGGUAAAGUUUGU----------------GAAAUGCAAACUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCU .(((.(((-((((((.((......)).).)))))))).))).((((((((((((----------------.....))))))))).((((.....))))...........)))........ ( -33.30) >pfo.1282 2551445 118 + 6438405 AUAGACUG-AUGGCCACUUUCACUGGUG-GUGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUCUUAGUUGAGAAAUUCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCU .(((.(((-((.((((((......))))-)).))))).))).(((.....((((((((((((......)))))............((((.....)))))))))))....)))........ ( -38.00) >consensus A_AGACUG_AUGACCUCUUUCACUGGUGAGCAUUCAAGCCAAGGUAAAAUUUGCGAGUU___________CAAACGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCU ...((((((((((((.((......)).))))))))).)))..(((((((((((((...................)))))))))).((((.....))))...........)))........ (-19.07 = -20.51 + 1.44)

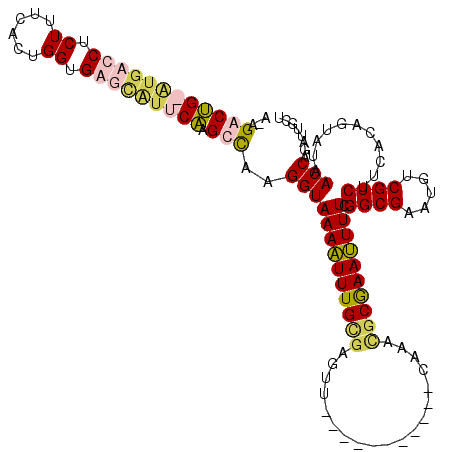
| Location | 724,022 – 724,131 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.38 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724022 109 + 6264404 AAGGUAAAAUUUGCGAGUU-----------CAAGCGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCUUGGGGUUAUAUGGUCAAGUGAAGAAGCGCAUACGGUGGAU .....((((((((((....-----------....))))))))))((.((.((((..(((((((.((((((((..(.....)..))))))))......)))))))...)))).)).))... ( -31.30) >pau.2632 1582397 109 - 6537648 AAGGUAAAAUUUGCGAGUU-----------CAAGCGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCUUGGGGUUAUAUGGUCAAGUGAAGAAGCGCAUACGGUGGAU .....((((((((((....-----------....))))))))))((.((.((((..(((((((.((((((((..(.....)..))))))))......)))))))...)))).)).))... ( -31.30) >pss.496 966697 104 + 6093698 AAGGUAAAGUUUGU----------------GAAAUGCAAACUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCUUGGGGUUAUAUGGUCAAGUGAAGAAGCGCAUACGGUGGAU .....(((((((((----------------.....)))))))))((.((.((((..(((((((.((((((((..(.....)..))))))))......)))))))...)))).)).))... ( -30.00) >pfo.1282 2551483 120 + 6438405 AAGGUAAAAUUUGUGAGUUCUCUUAGUUGAGAAAUUCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCUUGGGGUUAUAUGGUCAAGUGAAGAAGCGCAUACGGUGGAU .....((((((((....(((((......)))))...))))))))((.((.((((..(((((((.((((((((..(.....)..))))))))......)))))))...)))).)).))... ( -32.60) >consensus AAGGUAAAAUUUGCGAGUU___________CAAACGCGAAUUUUCGGCGAAUGUCGUCUUCACAGUAUAACCAGAUUGCUUGGGGUUAUAUGGUCAAGUGAAGAAGCGCAUACGGUGGAU .....((((((((((...................))))))))))((.((.((((..(((((((.((((((((..(.....)..))))))))......)))))))...)))).)).))... (-27.70 = -27.38 + -0.31)

| Location | 724,022 – 724,131 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -16.71 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724022 109 - 6264404 AUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCGCUUG-----------AACUCGCAAAUUUUACCUU ..........((((..(((((((.((((((.((...........)).))))))....)))))))...((..(((.((....)))))..))-----------....))))........... ( -21.70) >pau.2632 1582397 109 + 6537648 AUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCGCUUG-----------AACUCGCAAAUUUUACCUU ..........((((..(((((((.((((((.((...........)).))))))....)))))))...((..(((.((....)))))..))-----------....))))........... ( -21.70) >pss.496 966697 104 - 6093698 AUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAGUUUGCAUUUC----------------ACAAACUUUACCUU ......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))...((((((((......----------------.))))))))..... ( -22.40) >pfo.1282 2551483 120 - 6438405 AUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGAAUUUCUCAACUAAGAGAACUCACAAAUUUUACCUU ......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).)).(((....)))...(((((......)))))................. ( -21.60) >consensus AUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCACUUC___________AACUCACAAAUUUUACCUU ......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))...((((((.(((...................))).))))))..... (-16.71 = -17.34 + 0.63)

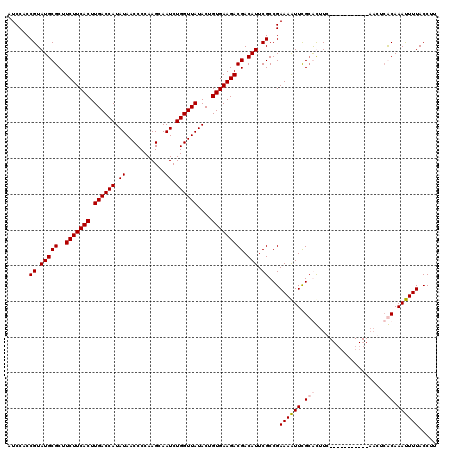
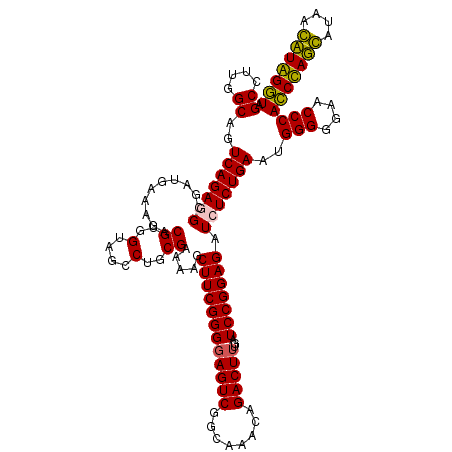
| Location | 724,131 – 724,251 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -43.88 |
| Energy contribution | -42.62 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724131 120 + 6264404 GCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUA ((....))..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....))))))). ( -45.70) >pau.2632 1582506 120 - 6537648 GCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUA ((....))..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....))))))). ( -45.70) >pss.496 966801 120 + 6093698 GCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUA (((((........)))))......(((((..(....)..)......(((((((.((((........))))....)))))))..))))....(((....)))(((((((....))))))). ( -41.20) >pfo.1282 2551603 120 + 6438405 GCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUA (((((........)))))......(((((..(....)..)......((((((((((((........)))))...)))))))..))))....(((....)))(((((((....))))))). ( -41.90) >consensus GCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAACAUAGGUA ((....))..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....))))))). (-43.88 = -42.62 + -1.25)

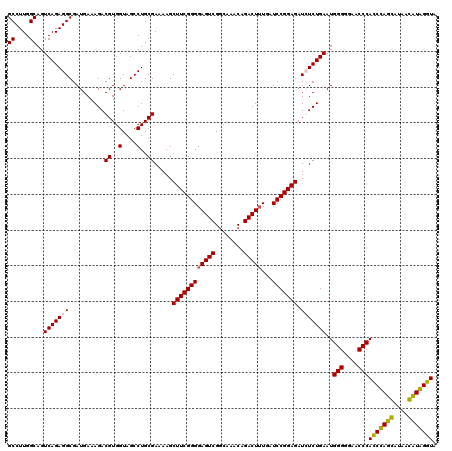
| Location | 724,171 – 724,291 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -44.67 |
| Energy contribution | -43.42 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.32 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724171 120 + 6264404 GAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGG ......((((((((((((........)))))...)))))))..(((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))). ( -46.60) >pau.2632 1582546 120 - 6537648 GAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGG ......((((((((((((........)))))...)))))))..(((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))). ( -46.60) >pss.496 966841 120 + 6093698 GAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGG ......(((((((.((((........))))....)))))))...((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......)))).. ( -41.00) >pfo.1282 2551643 120 + 6438405 GAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGG ......((((((((((((........)))))...)))))))...((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......)))).. ( -41.70) >consensus GAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAACAUAGGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGG ......((((((((((((........)))))...))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......)))))) (-44.67 = -43.42 + -1.25)

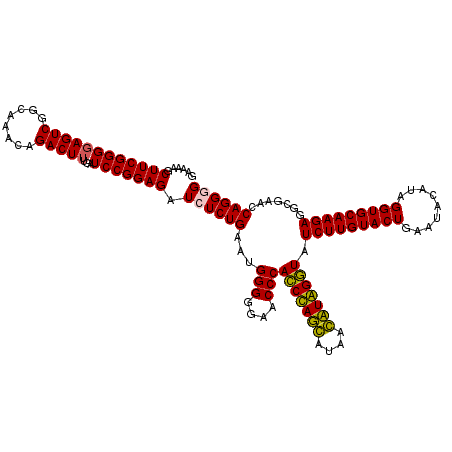
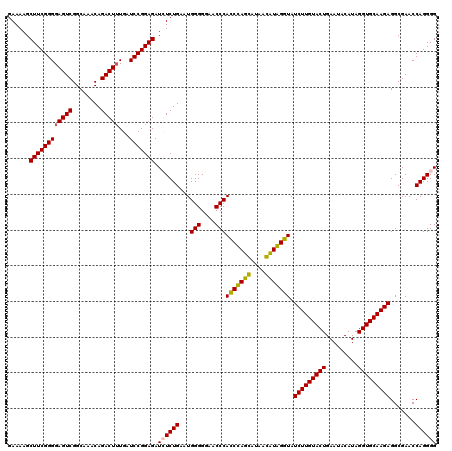
| Location | 724,211 – 724,331 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -41.25 |
| Energy contribution | -39.25 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.67 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 724211 120 + 6264404 GAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACC (((.(((....(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..))...))).))).... ( -40.20) >pau.2632 1582586 120 - 6537648 GAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACC (((.(((....(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..))...))).))).... ( -40.20) >pss.496 966881 120 + 6093698 GAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACC ((((((.....(((....)))(((((((....))))))).(((((((((.........))))))))))))...((.((((.(((..........))).))))..)).......))).... ( -38.40) >pfo.1282 2551683 120 + 6438405 GAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACC ((((((.....(((....)))(((((((....))))))).(((((((((.........))))))))))))...((.((((.(((..........))).))))..)).......))).... ( -38.40) >consensus GAUCUCUGAAUGGGGGAACCCACCCAGCAUAACAUAGGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACC (((.(((....(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..))...))).))).... (-41.25 = -39.25 + -2.00)

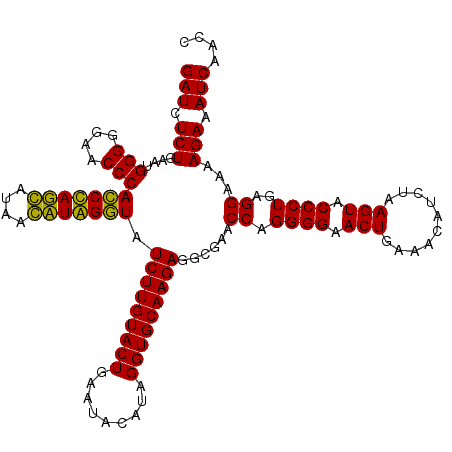
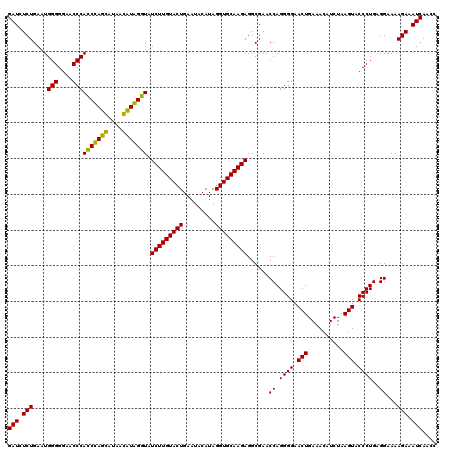
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:01 2007