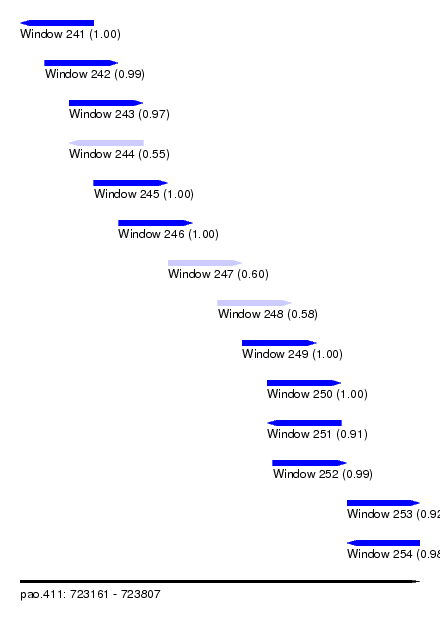
| Sequence ID | pao.411 |
|---|---|
| Location | 723,161 – 723,807 |
| Length | 646 |
| Max. P | 0.999963 |

| Location | 723,161 – 723,280 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.05 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -35.27 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723161 119 - 6264404 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACC-CGAGGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..((((((((-...))))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -36.70) >pau.2632 1581536 119 + 6537648 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACC-CGAGGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..((((((((-...))))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -36.70) >pss.496 965752 120 - 6093698 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -34.60) >ppk.94 172437 120 - 6181863 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -34.60) >pel.69 116557 120 - 5888780 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -34.60) >pfo.1282 2550595 120 - 6438405 UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUUACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))). ( -34.60) >consensus UCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGACA ................(((((.....)))))..((((((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))). (-35.27 = -35.27 + 0.00)

| Location | 723,201 – 723,320 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -46.07 |
| Consensus MFE | -44.43 |
| Energy contribution | -44.43 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
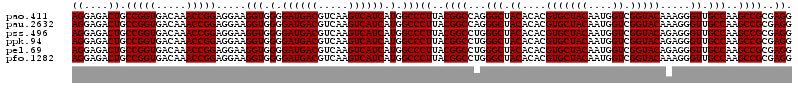
>pao.411 723201 119 + 6264404 CCUUGUCCUUAGUUACCAGCACCUCG-GGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACG ....((((.......(((.(.((((.-(((.((((.(((....))).))))(....)..))).))))..).))).((((((.....)))))).(((((.....)))))))))........ ( -48.00) >pau.2632 1581576 119 - 6537648 CCUUGUCCUUAGUUACCAGCACCUCG-GGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACG ....((((.......(((.(.((((.-(((.((((.(((....))).))))(....)..))).))))..).))).((((((.....)))))).(((((.....)))))))))........ ( -48.00) >pss.496 965792 120 + 6093698 CCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACG ((((.(((((((...(((.((.....)).))).....))))))).)...(((((.....))))))))...(((..((((((.....)))))).((((((.........)))))))))... ( -45.10) >ppk.94 172477 120 + 6181863 CCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACG ((((.(((((((...(((.((.....)).))).....))))))).)...(((((.....))))))))...(((..((((((.....)))))).((((((.........)))))))))... ( -45.10) >pel.69 116597 120 + 5888780 CCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACG ((((.(((((((...(((.((.....)).))).....))))))).)...(((((.....))))))))...(((..((((((.....)))))).((((((.........)))))))))... ( -45.10) >pfo.1282 2550635 120 + 6438405 CCUUGUCCUUAGUUACCAGCACGUAAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACG ((((.(((((((...(((.((.....)).))).....))))))).)...(((((.....))))))))...(((..((((((.....)))))).((((((.........)))))))))... ( -45.10) >consensus CCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACG (((..(((((.((((((..(((......)))((((.(((....))).))))))))))......)))))....)))((((((.....)))))).((((((.........))))))...... (-44.43 = -44.43 + -0.00)

| Location | 723,240 – 723,360 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.06 |
| Mean single sequence MFE | -45.07 |
| Consensus MFE | -44.80 |
| Energy contribution | -44.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
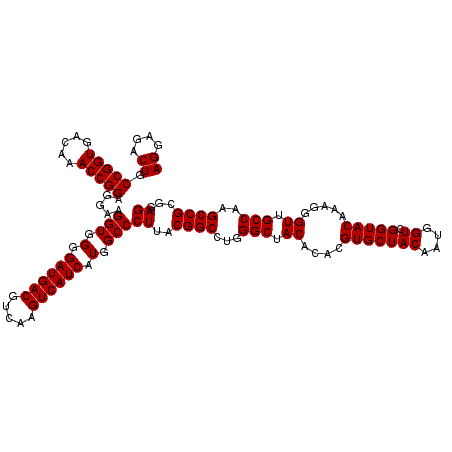
>pao.411 723240 120 + 6264404 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((...((((((.....))))))..(((((((.((((((.(((.((....)))))....)))))).....))))))))))...((....)) ( -45.60) >pau.2632 1581615 120 - 6537648 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((...((((((.....))))))..(((((((.((((((.(((.((....)))))....)))))).....))))))))))...((....)) ( -45.60) >pss.496 965832 120 + 6093698 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((.(.((((((.....)))))).).)))((..((((...(((.((....(((((((....)).))))).....)).)))..))))..)). ( -44.80) >ppk.94 172517 120 + 6181863 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((.(.((((((.....)))))).).)))((..((((...(((.((....(((((((....)).))))).....)).)))..))))..)). ( -44.80) >pel.69 116637 120 + 5888780 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((.(.((((((.....)))))).).)))((..((((...(((.((....(((((((....)).))))).....)).)))..))))..)). ( -44.80) >pfo.1282 2550675 120 + 6438405 AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((.(.((((((.....)))))).).)))((..((((...(((.((....(((((((....)).))))).....)).)))..))))..)). ( -44.80) >consensus AGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGG ((....)).(((((.....))))).....(((.(.((((((.....)))))).).)))((..((((...(((.((....(((((((....)).))))).....)).)))..))))..)). (-44.80 = -44.80 + 0.00)
| Location | 723,240 – 723,360 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.06 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -36.27 |
| Energy contribution | -36.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723240 120 - 6264404 CCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCUGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((.(((.((.(..(((..(((...)))..)))...).)).)))..)))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -36.50) >pau.2632 1581615 120 + 6537648 CCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCUGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((.(((.((.(..(((..(((...)))..)))...).)).)))..)))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -36.50) >pss.496 965832 120 - 6093698 CCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -38.40) >ppk.94 172517 120 - 6181863 CCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -38.40) >pel.69 116637 120 - 5888780 CCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -38.40) >pfo.1282 2550675 120 - 6438405 CCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((((((.((.(..(((..(((...)))..)))...).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))........ ( -37.50) >consensus CCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCU (((..(((((((((.((.(..(((..(((...)))..)))...).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))........ (-36.27 = -36.60 + 0.33)

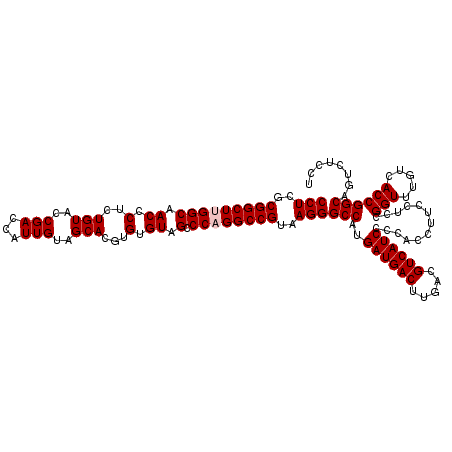
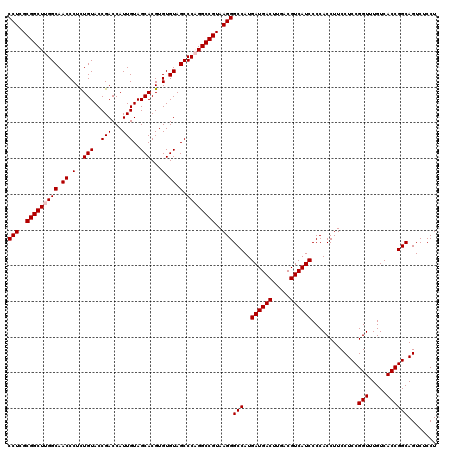
| Location | 723,280 – 723,400 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -42.83 |
| Energy contribution | -42.58 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723280 120 + 6264404 CGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((.(((((.....)))))(((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))).))).)). ( -45.20) >pau.2632 1581655 120 - 6537648 CGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((.(((((.....)))))(((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))).))).)). ( -45.20) >pss.496 965872 120 + 6093698 CGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((..((((.....))))((((((((....(.....)...(((((((...((((...((...(((....)))...))...)))).....)))))))))))))))))).)). ( -42.90) >ppk.94 172557 120 + 6181863 CGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((..((((.....))))((((((((....(.....)...(((((((...((((...((...(((....)))...))...)))).....)))))))))))))))))).)). ( -42.90) >pel.69 116677 120 + 5888780 CGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((..((((.....))))((((((((....(.....)...(((((((...((((...((...(((....)))...))...)))).....)))))))))))))))))).)). ( -42.90) >pfo.1282 2550715 120 + 6438405 CGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((..((((.....))))((((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))))))).)). ( -43.60) >consensus CGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCA ......((.(((..((((.....)))).(((((((....(.....)...(((((((.....(((((((...(((....)))...)).)))))......)))))))))))))).))).)). (-42.83 = -42.58 + -0.25)
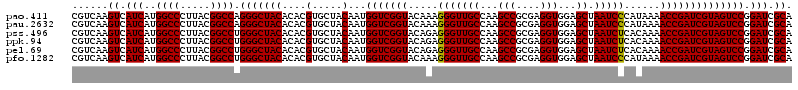
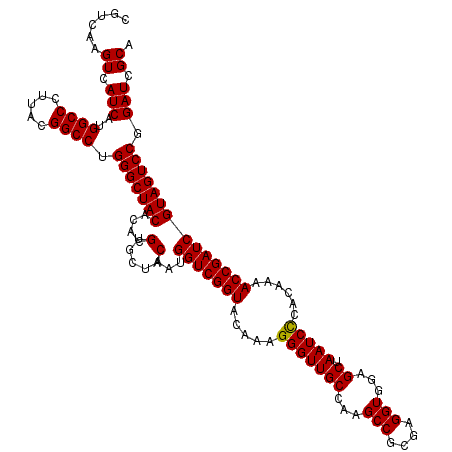
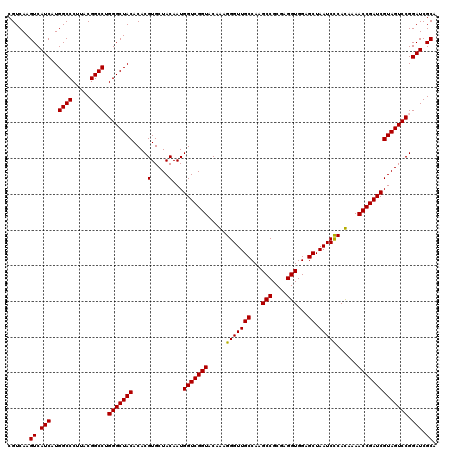
| Location | 723,320 – 723,440 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -45.55 |
| Consensus MFE | -45.00 |
| Energy contribution | -44.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723320 120 + 6264404 UGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.90) >pau.2632 1581695 120 - 6537648 UGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.90) >pss.496 965912 120 + 6093698 UGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((...((((...((...(((....)))...))...)))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.20) >ppk.94 172597 120 + 6181863 UGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((...((((...((...(((....)))...))...)))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.20) >pel.69 116717 120 + 5888780 UGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((...((((...((...(((....)))...))...)))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.20) >pfo.1282 2550755 120 + 6438405 UGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).))))).. ( -45.90) >consensus UGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAU ((((((.(((((((((.....(((((((...(((....)))...)).)))))......)))))))))..(((((..(((((((........))))))).....)))))...).))))).. (-45.00 = -44.75 + -0.25)
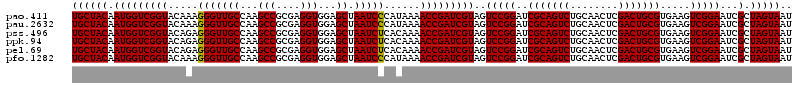
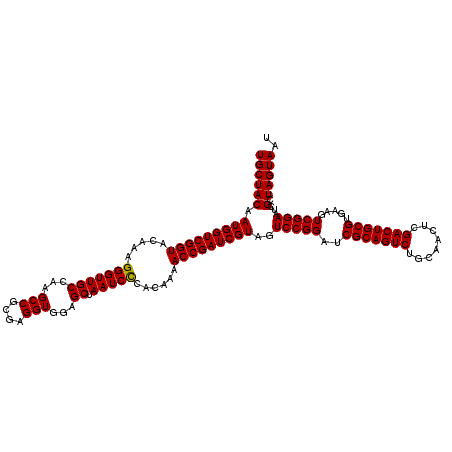
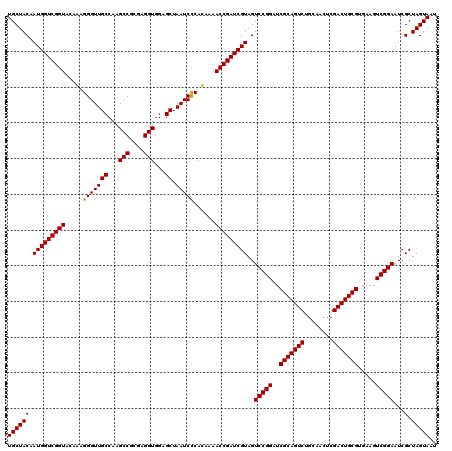
| Location | 723,400 – 723,520 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -40.51 |
| Consensus MFE | -40.55 |
| Energy contribution | -39.83 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.61 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723400 120 + 6264404 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGUGAAUCAGAAUGUCACGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCA ....(((((((.(((.(((((..((.((.((((.(....).)))).)).))...))))((((..........(((((..((....))..)))))...))))...).)))))))))).... ( -38.92) >pau.2632 1581775 120 - 6537648 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGUGAAUCAGAAUGUCACGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCA ....(((((((.(((.(((((..((.((.((((.(....).)))).)).))...))))((((..........(((((..((....))..)))))...))))...).)))))))))).... ( -38.92) >pss.496 965992 120 + 6093698 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCA ...(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..))))))))))))))... ( -41.90) >ppk.94 172677 120 + 6181863 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCA ...(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..))))))))))))))... ( -41.90) >pel.69 116797 120 + 5888780 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCAAAUCAGAAUGUUGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCA ...(((((((((..(.((((.....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..)))))......)))).)..)))))))))... ( -39.50) >pfo.1282 2550835 120 + 6438405 GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCA ...(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..))))))))))))))... ( -41.90) >consensus GUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCA ....((((((((..(.((((.....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..)))))......)))).)..)))))))).... (-40.55 = -39.83 + -0.72)

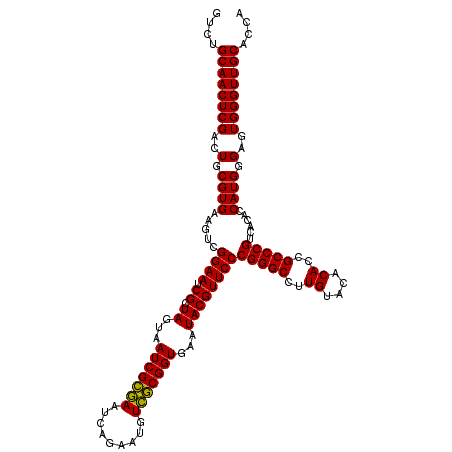
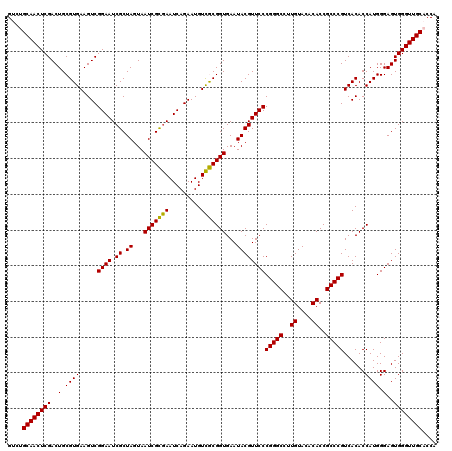
| Location | 723,480 – 723,600 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -40.29 |
| Energy contribution | -41.85 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723480 120 + 6264404 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG .....(((..(((((......))))).)))(((((((.....))))))).((((..((....)).))))(((((((..(((....)))((((((.......)))))).....))))))). ( -43.40) >pau.2632 1581855 120 - 6537648 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG .....(((..(((((......))))).)))(((((((.....))))))).((((..((....)).))))(((((((..(((....)))((((((.......)))))).....))))))). ( -43.40) >pss.496 966072 120 + 6093698 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG ......(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))..... ( -41.30) >ppk.94 172757 120 + 6181863 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG ......(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))..... ( -41.30) >pel.69 116877 120 + 5888780 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG ......(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))..... ( -42.00) >pfo.1282 2550915 120 + 6438405 GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG ......(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))..... ( -41.30) >consensus GUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCG ......(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))..... (-40.29 = -41.85 + 1.56)


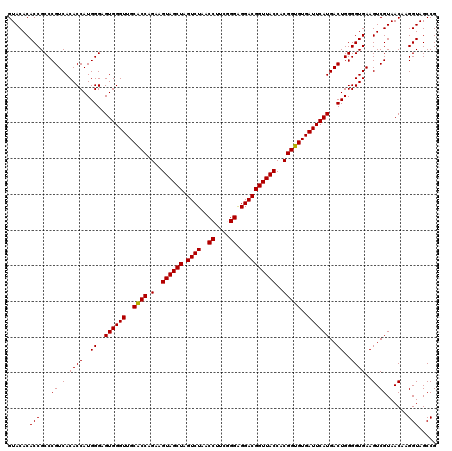
| Location | 723,520 – 723,640 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -50.12 |
| Consensus MFE | -46.55 |
| Energy contribution | -48.55 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.76 |
| SVM RNA-class probability | 0.999947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723520 120 + 6264404 GAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAU ((.((((((.((((..((....)).))))))))))...((((((((((((((((.......))))))........(((((((((....))))))))))))))).))))....))...... ( -52.00) >pau.2632 1581895 120 - 6537648 GAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAU ((.((((((.((((..((....)).))))))))))...((((((((((((((((.......))))))........(((((((((....))))))))))))))).))))....))...... ( -52.00) >pss.496 966112 120 + 6093698 GAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC ((.((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))....))...... ( -49.00) >ppk.94 172797 120 + 6181863 GAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC ((.((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))....))...... ( -49.00) >pel.69 116917 120 + 5888780 GAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC ((.((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))....))...... ( -49.70) >pfo.1282 2550955 120 + 6438405 GAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC ((.((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))....))...... ( -49.00) >consensus GAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC ((.((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))....))...... (-46.55 = -48.55 + 2.00)

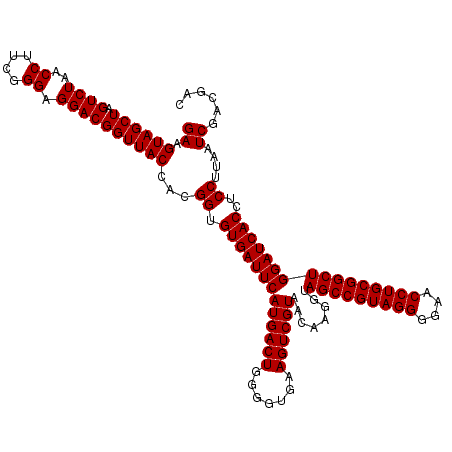
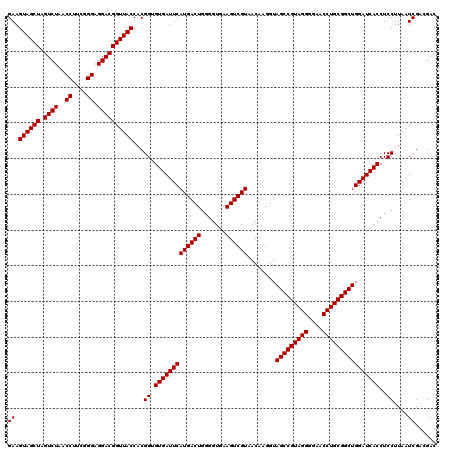
| Location | 723,560 – 723,680 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -45.65 |
| Consensus MFE | -42.08 |
| Energy contribution | -42.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723560 120 + 6264404 AGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUC ((..((((.((...(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........)))))...........)).))))..))...... ( -42.30) >pau.2632 1581935 120 - 6537648 AGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUC ((..((((.((...(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........)))))...........)).))))..))...... ( -42.30) >pss.496 966152 119 + 6093698 UGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCGCCAUAAGCACCCACACGAAUUGCUUGAUUC .(..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).)))))).))))..)....... ( -46.40) >ppk.94 172837 120 + 6181863 UGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUC .(..((((.((..((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).)))))).))))..)....... ( -49.40) >pel.69 116957 120 + 5888780 UGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUC .(..((((.((..((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).)))))).))))..)....... ( -49.40) >pfo.1282 2550995 120 + 6438405 UGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCUCCAUAAGUUCCCACACGAAUUGCUUGAUUC .(..((((.((..((((((((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........)))))))))).))))..)....... ( -44.10) >consensus UGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUC .(..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))).)))))).))))..)....... (-42.08 = -42.38 + 0.31)
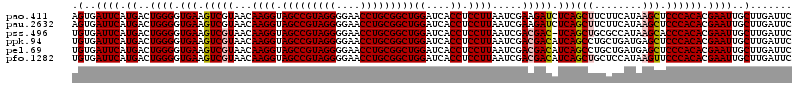
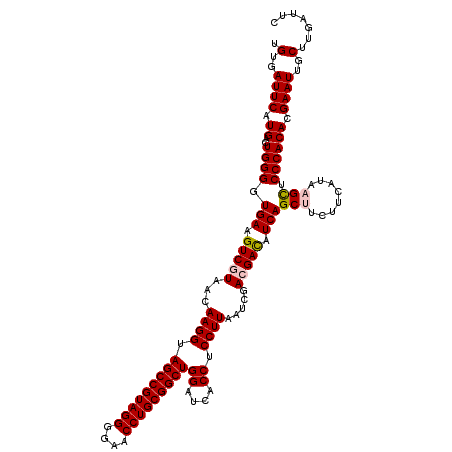

| Location | 723,560 – 723,680 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -35.15 |
| Energy contribution | -35.93 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723560 120 - 6264404 GAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAGAUCUUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACU ......((..((((((((((((((((......)))))..((...(((..(((((.((....))(((((.(((....))).))))).)))))...)))..))..))))..)))))))..)) ( -36.90) >pau.2632 1581935 120 + 6537648 GAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAGAUCUUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACU ......((..((((((((((((((((......)))))..((...(((..(((((.((....))(((((.(((....))).))))).)))))...)))..))..))))..)))))))..)) ( -36.90) >pss.496 966152 119 - 6093698 GAAUCAAGCAAUUCGUGUGGGUGCUUAUGGCGCAGCUGA-GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACA .......(..(((((((.(((((.....(((...)))((-(((((....(((((.((....))(((((.(((....))).))))).)))))..))))))))))))....)))))))..). ( -39.70) >ppk.94 172837 120 - 6181863 GAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGCAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACA .......(..(((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))))..). ( -42.30) >pel.69 116957 120 - 5888780 GAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGCAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACA .......(..(((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))))..). ( -42.30) >pfo.1282 2550995 120 - 6438405 GAAUCAAGCAAUUCGUGUGGGAACUUAUGGAGCAGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACA ((.((((((..((((((.(....).))))))...)))((.(((((....(((((.((....))(((((.(((....))).))))).)))))..))))).))..........))).))... ( -36.10) >consensus GAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCACA .......(..(((((((((((((((........))))((.(((((....(((((.((....))(((((.(((....))).))))).)))))..))))).))..))))..)))))))..). (-35.15 = -35.93 + 0.78)

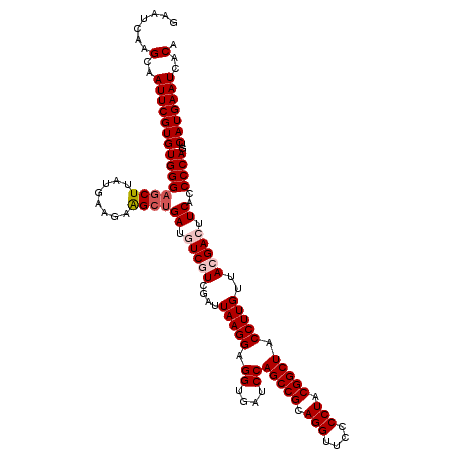
| Location | 723,569 – 723,689 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -37.71 |
| Energy contribution | -38.02 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723569 120 + 6264404 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCACUGGUUAG ((((..((((((.((....))....((((((((((....))))))))))..))))).....(((((((((........)))))..((((.............))))))))..)..)))). ( -40.62) >pau.2632 1581944 120 - 6537648 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCACUGGUUAG ((((..((((((.((....))....((((((((((....))))))))))..))))).....(((((((((........)))))..((((.............))))))))..)..)))). ( -40.62) >pss.496 966161 115 + 6093698 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCGCCAUAAGCACCCACACGAAUUGCUUGAUUCAUUGA---- ....((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).))))...(((((....))))).....---- ( -42.10) >ppk.94 172846 116 + 6181863 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUCAUUGU---- ....((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).))))...(((((....))))).....---- ( -45.10) >pel.69 116966 116 + 5888780 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUCAGAUG---- ....((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).))))...(((((....))))).....---- ( -45.10) >pfo.1282 2551004 116 + 6438405 UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCUCCAUAAGUUCCCACACGAAUUGCUUGAUUCAUUGA---- ....((((((((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))))))))...(((((....))))).....---- ( -39.80) >consensus UGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCAUUGG____ ....((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))).))))...(((((....)))))......... (-37.71 = -38.02 + 0.31)
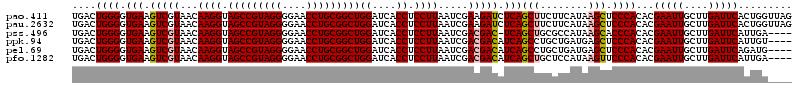
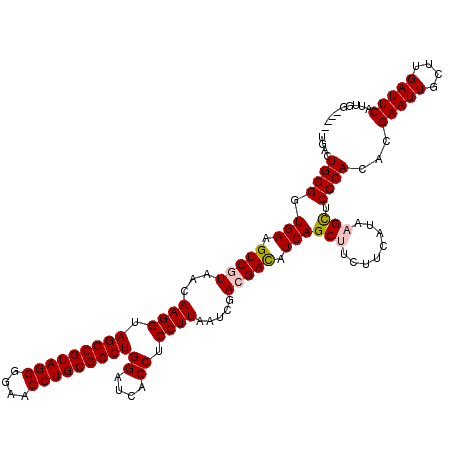
| Location | 723,689 – 723,807 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -47.45 |
| Consensus MFE | -38.35 |
| Energy contribution | -39.85 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723689 118 + 6264404 ACGAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUUGUUGGUGUGCUGCG--UGAUCCGAUACGGGGCCA .((((((((((((((((((.........))))((((.((.(((((.((((.(((((((((.......)))))..))))))))..))))))).))))))))--.)))))))).))...... ( -49.50) >pau.2632 1582064 118 - 6537648 ACGAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUUGUUGGUGUGCUGCG--UGAUCCGAUACGGGGCCA .((((((((((((((((((.........))))((((.((.(((((.((((.(((((((((.......)))))..))))))))..))))))).))))))))--.)))))))).))...... ( -49.50) >pss.496 966340 120 + 6093698 GAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUUUUGUGUGGGAAACGCCUGUAGAAAUACGGGGCCA ..((((((((.....))))))))(((..((.(.(((((.......))))).).))(((((.......)))))....(((((((....)).))))).....((((((....))))))))). ( -45.40) >pfo.1282 2551141 120 + 6438405 GAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUUUUGUGUGGGAAACGCCUGUAGAAAUACGGGGCCA ..((((((((.....))))))))(((..((.(.(((((.......))))).).))(((((.......)))))....(((((((....)).))))).....((((((....))))))))). ( -45.40) >consensus AAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUUGUGGGUGGGAAACG__UGAACAAAUACGGGGCCA .....(((.((((((((((.........)))).(((((.((((...((((.(((((((((.......)))))..)))))))).)))).))))).................)))))).))) (-38.35 = -39.85 + 1.50)

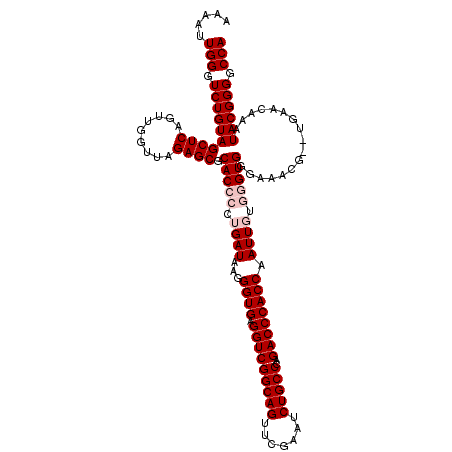
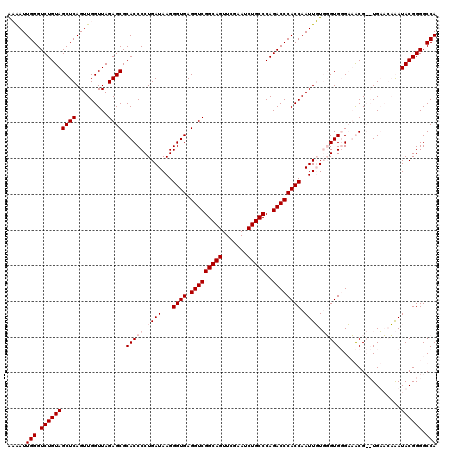
| Location | 723,689 – 723,807 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 723689 118 - 6264404 UGGCCCCGUAUCGGAUCA--CGCAGCACACCAACAAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUCGU ..((..(((........)--))..))..((((.....))))((((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))))...... ( -41.40) >pau.2632 1582064 118 + 6537648 UGGCCCCGUAUCGGAUCA--CGCAGCACACCAACAAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUCGU ..((..(((........)--))..))..((((.....))))((((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))))...... ( -41.40) >pss.496 966340 120 - 6093698 UGGCCCCGUAUUUCUACAGGCGUUUCCCACACAAAAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUC ..(((..(((....))).))).....(((........)))(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))..... ( -41.90) >pfo.1282 2551141 120 - 6438405 UGGCCCCGUAUUUCUACAGGCGUUUCCCACACAAAAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUC ..(((..(((....))).))).....(((........)))(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))..... ( -41.90) >consensus UGGCCCCGUAUCGCAACA__CGCAGCACACAAAAAAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUCGC ........................................(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))..... (-36.10 = -36.10 + 0.00)

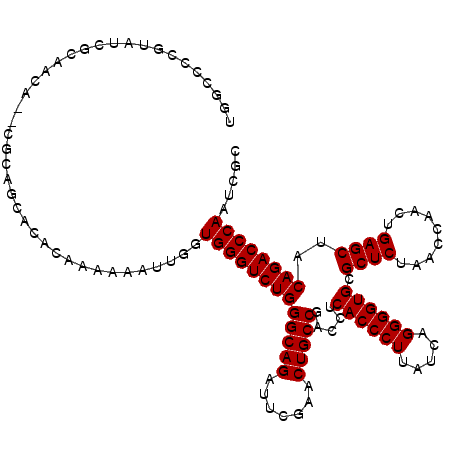
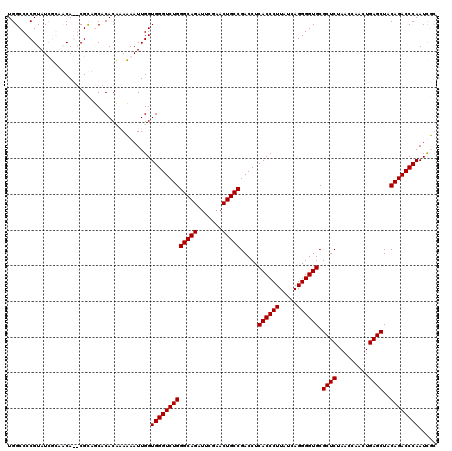
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:07:50 2007