
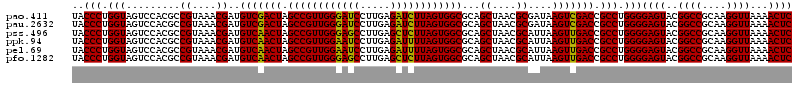
| Sequence ID | pao.411 |
|---|---|
| Location | 722,201 – 723,001 |
| Length | 800 |
| Max. P | 0.999979 |

| Location | 722,201 – 722,321 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -39.34 |
| Energy contribution | -41.45 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
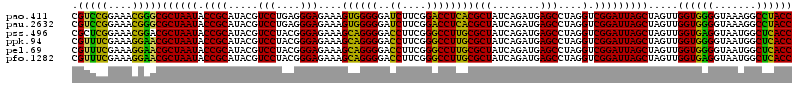
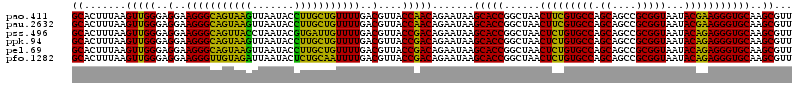
>pao.411 722201 120 + 6264404 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAG ........((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).((((.(((...((...((((((..((....)))))))).)).))))))).. ( -45.00) >pau.2632 1580576 120 - 6537648 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAG ........((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).((((.(((...((...((((((..((....)))))))).)).))))))).. ( -45.00) >pss.496 964792 120 + 6093698 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAG ....(((.((((.(((......)))))))((((....((.((..((....))..)).))...)))).))).(((....)))((.(((.((((.((....))))))..))).))....... ( -41.10) >ppk.94 171477 120 + 6181863 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAG ....(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))....... ( -42.82) >pel.69 115597 120 + 5888780 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAG ....(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))....... ( -42.82) >pfo.1282 2549635 120 + 6438405 GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAG ....(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))....... ( -42.82) >consensus GUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAG .((((((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))....((((((..((....))))))))....)))...... (-39.34 = -41.45 + 2.12)

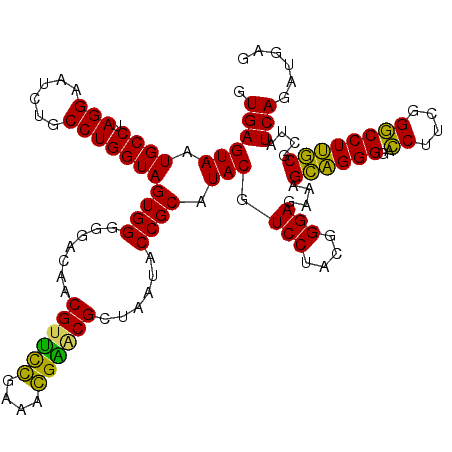
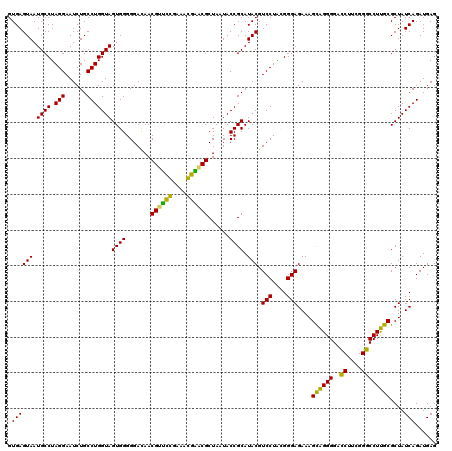
| Location | 722,241 – 722,361 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -43.42 |
| Energy contribution | -44.87 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722241 120 + 6264404 CGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACC .(((((....)))))((((((.(((....((((.(((...((...((((((..((....)))))))).)).))))))).((....))))))))))).....((((((.......)))))) ( -47.90) >pau.2632 1580616 120 - 6537648 CGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACC .(((((....)))))((((((.(((....((((.(((...((...((((((..((....)))))))).)).))))))).((....))))))))))).....((((((.......)))))) ( -47.90) >pss.496 964832 120 + 6093698 CGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACC .(..((....))..)((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).))))))))).....((((((.......)))))) ( -44.20) >ppk.94 171517 120 + 6181863 CGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACC .(((((....)))))((((.((((.(((...(((....)))....((((((..((....))))))))((((((.(((........))).)).))))......))).)))).))))..... ( -45.50) >pel.69 115637 120 + 5888780 CGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACC .(((((....)))))((((.((((.(((...(((....)))....((((((..((....))))))))((((((.(((........))).)).))))......))).)))).))))..... ( -45.50) >pfo.1282 2549675 120 + 6438405 CGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACC .(((((....)))))((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).))))))))).....((((((.......)))))) ( -46.00) >consensus CGUUCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACC .(((((....)))))((((((.((((.....(((....)))....((((((..((....))))))))(((........)))....).))))))))).....((((((.......)))))) (-43.42 = -44.87 + 1.45)
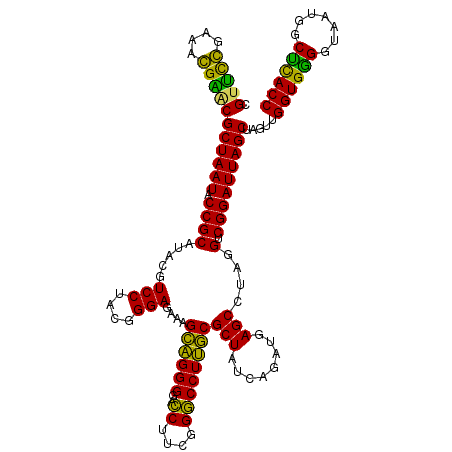
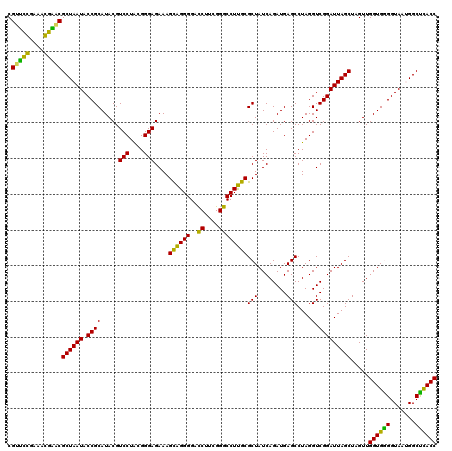
| Location | 722,281 – 722,401 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -43.16 |
| Energy contribution | -44.17 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722281 120 + 6264404 AGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .((.((((((((.((....)))))).)))).)).(((.(..((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....)))).))).... ( -45.20) >pau.2632 1580656 120 - 6537648 AGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .((.((((((((.((....)))))).)))).)).(((.(..((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....)))).))).... ( -45.20) >pss.496 964872 120 + 6093698 AGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .....((((((..((....))))))))(((((((.......((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....))))))).))). ( -45.30) >ppk.94 171557 120 + 6181863 AGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .....((((((..((....))))))))(((((((.......((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....))))))).))). ( -44.40) >pel.69 115677 120 + 5888780 AGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .....((((((..((....))))))))(((((((.......((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....))))))).))). ( -44.40) >pfo.1282 2549715 120 + 6438405 AGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .....((((((..((....))))))))(((((((.......((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....))))))).))). ( -45.30) >consensus AGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUC .((..((((((..((....))))))))..((((...(.((.((((..((((((.(((..((((((((.......)))))))))))...))))))...)))).)).)...)))).)).... (-43.16 = -44.17 + 1.00)

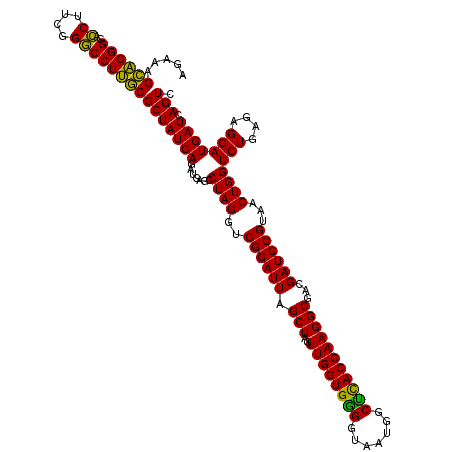
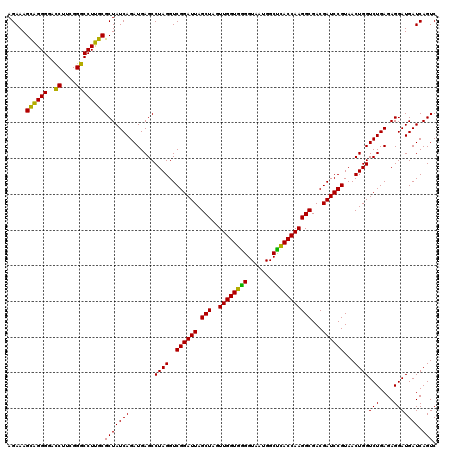
| Location | 722,321 – 722,441 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -44.90 |
| Energy contribution | -46.73 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722321 120 + 6264404 CCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -46.90) >pau.2632 1580696 120 - 6537648 CCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -46.90) >pss.496 964912 120 + 6093698 CCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -47.10) >ppk.94 171597 120 + 6181863 CCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -46.20) >pel.69 115717 120 + 5888780 CCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -46.20) >pfo.1282 2549755 120 + 6438405 CCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. ( -47.10) >consensus CCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGC ........(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....)))).. (-44.90 = -46.73 + 1.83)

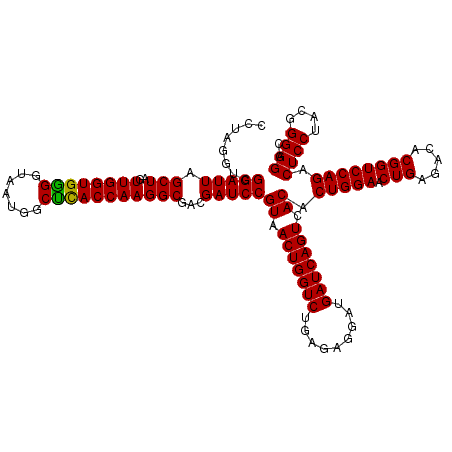
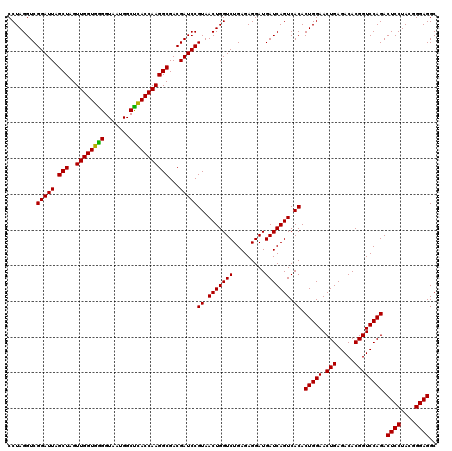
| Location | 722,361 – 722,481 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -38.20 |
| Energy contribution | -40.70 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722361 120 + 6264404 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >pau.2632 1580736 120 - 6537648 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >pss.496 964952 120 + 6093698 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >ppk.94 171637 120 + 6181863 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >pel.69 115757 120 + 5888780 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >pfo.1282 2549795 120 + 6438405 AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ...(((......))).....((((....))))((((.(((...(((((.(((.....)))))))).((((....))))....(((((.....))))))))...((((....)))))))). ( -40.70) >consensus AAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCC ..(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).))).....(((...(((((....))))).))) (-38.20 = -40.70 + 2.50)

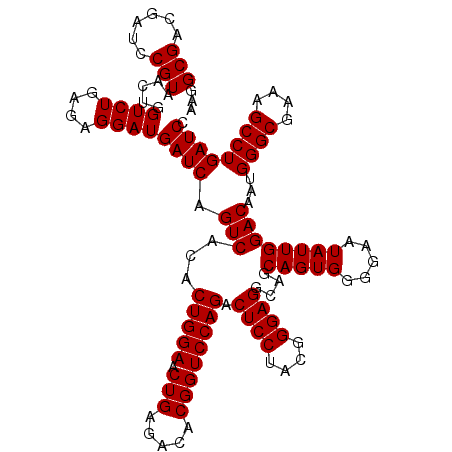
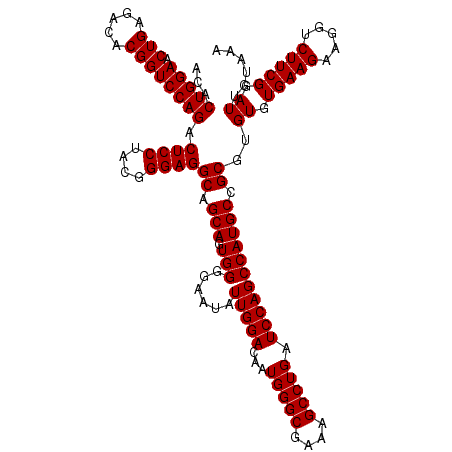
| Location | 722,401 – 722,521 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -40.00 |
| Energy contribution | -42.50 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722401 120 + 6264404 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >pau.2632 1580776 120 - 6537648 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >pss.496 964992 120 + 6093698 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >ppk.94 171677 120 + 6181863 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >pel.69 115797 120 + 5888780 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >pfo.1282 2549835 120 + 6438405 ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... ( -42.50) >consensus ACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAA ...(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))...... (-40.00 = -42.50 + 2.50)

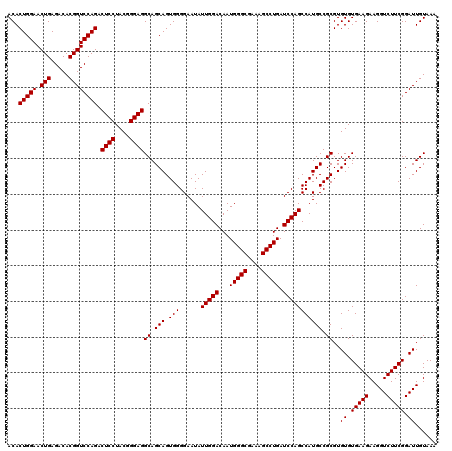
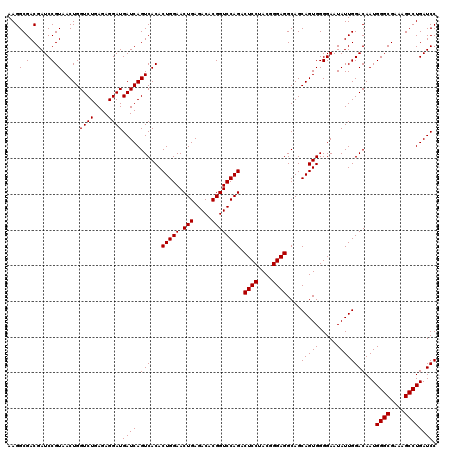
| Location | 722,401 – 722,521 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722401 120 - 6264404 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >pau.2632 1580776 120 + 6537648 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >pss.496 964992 120 - 6093698 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >ppk.94 171677 120 - 6181863 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >pel.69 115797 120 - 5888780 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >pfo.1282 2549835 120 - 6438405 UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) ( -35.20) >consensus UUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGU ..........((((.....))))..(((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).)))) (-35.20 = -35.20 + -0.00)

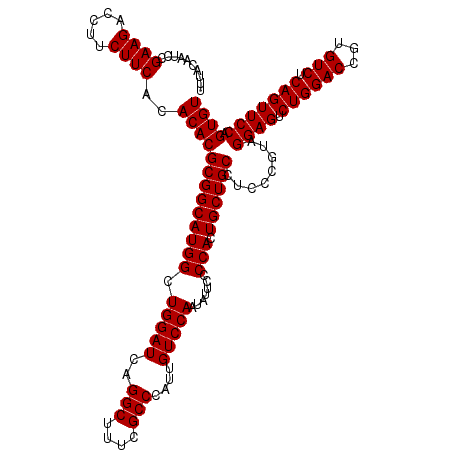
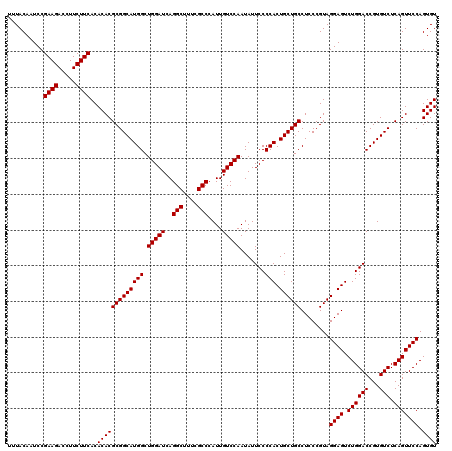
| Location | 722,481 – 722,601 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -38.59 |
| Energy contribution | -39.48 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722481 120 + 6264404 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -40.30) >pau.2632 1580856 120 - 6537648 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -40.30) >pss.496 965072 120 + 6093698 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -37.80) >ppk.94 171757 120 + 6181863 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -40.00) >pel.69 115877 120 + 5888780 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -40.00) >pfo.1282 2549915 120 + 6438405 AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... ( -38.50) >consensus AGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACU ......((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))..... (-38.59 = -39.48 + 0.90)

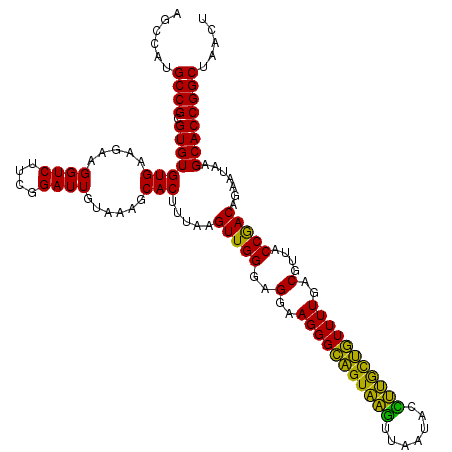
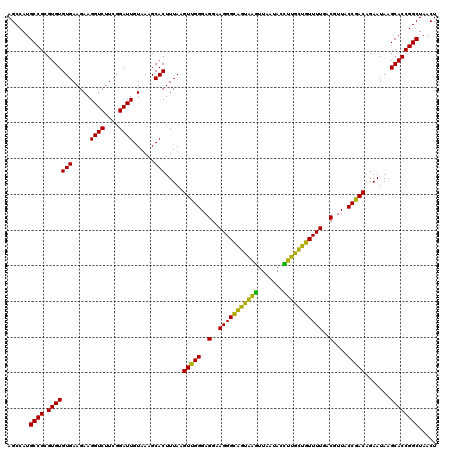
| Location | 722,521 – 722,641 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -43.58 |
| Energy contribution | -40.58 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.97 |
| Structure conservation index | 1.07 |
| SVM decision value | 5.22 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722521 120 + 6264404 GCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUGCCAGCAGCCGCGGUAAUACGAAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -40.80) >pau.2632 1580896 120 - 6537648 GCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUGCCAGCAGCCGCGGUAAUACGAAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -40.80) >pss.496 965112 120 + 6093698 GCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -39.20) >ppk.94 171797 120 + 6181863 GCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -41.40) >pel.69 115917 120 + 5888780 GCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -41.40) >pfo.1282 2549955 120 + 6438405 GCACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... ( -39.90) >consensus GCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUU ((.......(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))..))... (-43.58 = -40.58 + -2.99)
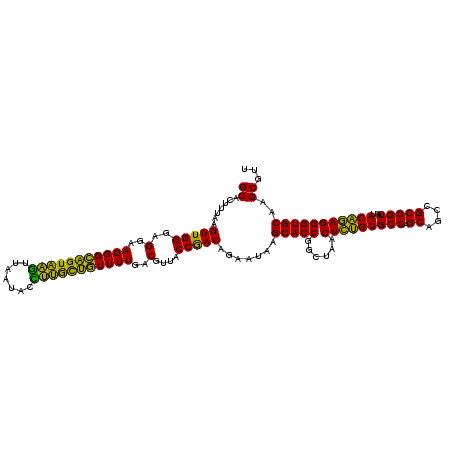
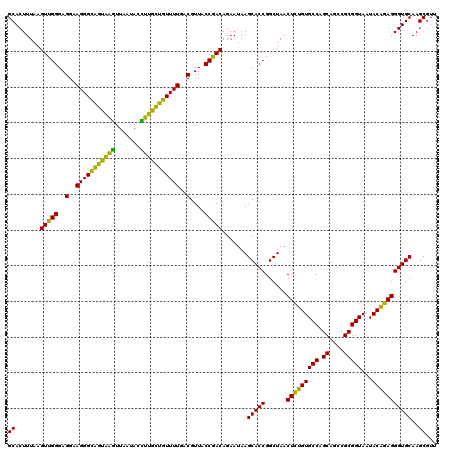
| Location | 722,641 – 722,761 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -41.60 |
| Energy contribution | -40.52 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.03 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722641 120 + 6264404 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCAGCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGU .((((......(((.((((.....)))).))).((((((((....(((((((((...((((.((......)).))))..))))))))).....)))))))).....)))).......... ( -42.00) >pau.2632 1581016 120 - 6537648 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCAGCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGU .((((......(((.((((.....)))).))).((((((((....(((((((((...((((.((......)).))))..))))))))).....)))))))).....)))).......... ( -42.00) >pss.496 965232 120 + 6093698 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGU .((((...((.(((.((((.....)))).))).((((((((((..(((.(((((...((((.((......)).))))..))))).)))...))))))))))))...)))).......... ( -33.30) >ppk.94 171917 120 + 6181863 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGU .((((......(((.((((.....)))).))).((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).....)))).......... ( -42.10) >pel.69 116037 120 + 5888780 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGU .((((...((.(((.((((.....)))).))).((((((((((..((((((((......((((((.....))))))....))))))))...))))))))))))...)))).......... ( -41.70) >pfo.1282 2550075 120 + 6438405 AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGU .((((...((.(((.((((.....)))).))).((((((((((..(((((((((...((((.((......)).))))..)))))))))...))))))))))))...)))).......... ( -41.90) >consensus AAUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUACGGUAGAGGGUGGU .((((......(((.((((.....)))).))).((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).....)))).......... (-41.60 = -40.52 + -1.08)

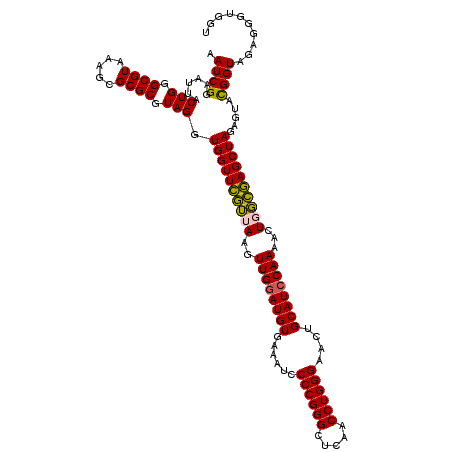
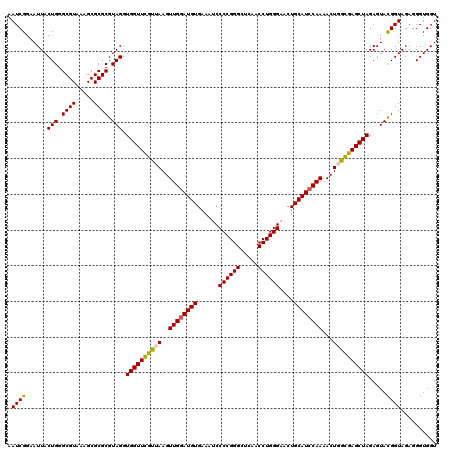
| Location | 722,681 – 722,801 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -34.02 |
| Energy contribution | -33.63 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722681 120 + 6264404 GCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .....(((((((((...((((.((......)).))))..)))))))))..((((((.((....)).)))))).............(((((((((.(((......)))....))))))))) ( -39.70) >pau.2632 1581056 120 - 6537648 GCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .....(((((((((...((((.((......)).))))..)))))))))..((((((.((....)).)))))).............(((((((((.(((......)))....))))))))) ( -39.70) >pss.496 965272 120 + 6093698 UUAAGUUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .................((((((..(((.(((..(((......)))....((((....)))).....))).)))..))).)))..(((((((((.(((......)))....))))))))) ( -32.50) >ppk.94 171957 120 + 6181863 UUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .....((((((((......((((((.....))))))....))))))))..((((....))))(.(((........))).).....(((((((((.(((......)))....))))))))) ( -35.10) >pel.69 116077 120 + 5888780 UUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA ...((((....((....))((((((.....))))))))))...((((..(((..(..((((.....)))).)..)))..))))..(((((((((.(((......)))....))))))))) ( -35.60) >pfo.1282 2550115 120 + 6438405 UUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .....(((((((((...((((.((......)).))))..)))))))))..(((.((.((....)).)).))).............(((((((((.(((......)))....))))))))) ( -35.50) >consensus UUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAA .....((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).))).............(((((((((.(((......)))....))))))))) (-34.02 = -33.63 + -0.39)

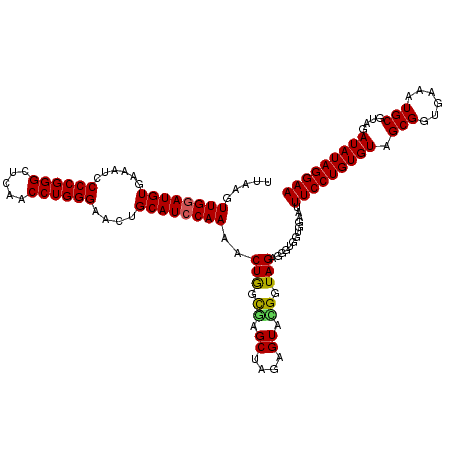
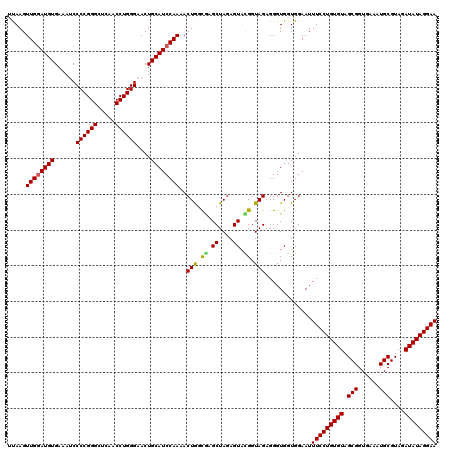
| Location | 722,721 – 722,841 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -41.29 |
| Energy contribution | -41.15 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.15 |
| SVM RNA-class probability | 0.999976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722721 120 + 6264404 GCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG .(((((....((((((.((....)).))))))(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))))).))...... ( -43.30) >pau.2632 1581096 120 - 6537648 GCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG .(((((....((((((.((....)).))))))(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))))).))...... ( -43.30) >pss.496 965312 120 + 6093698 GCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG ..........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))). ( -42.30) >ppk.94 171997 120 + 6181863 GCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG ..........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).)))). ( -43.80) >pel.69 116117 120 + 5888780 GCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG ..........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))). ( -42.30) >pfo.1282 2550155 120 + 6438405 GCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG ..........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))). ( -42.30) >consensus GCAUCCAAAACUGGCGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUG ..........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).)))). (-41.29 = -41.15 + -0.14)
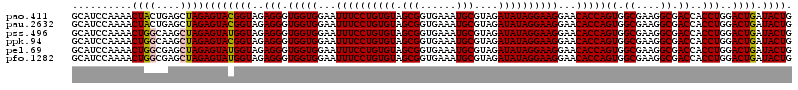
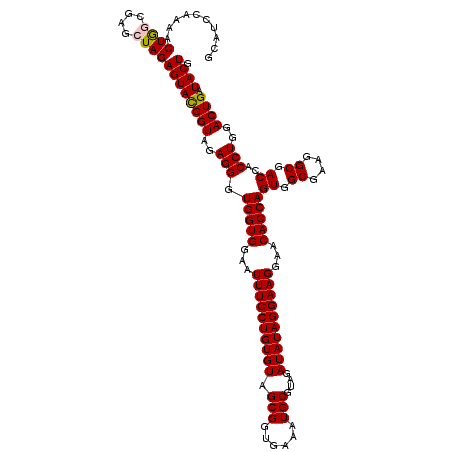
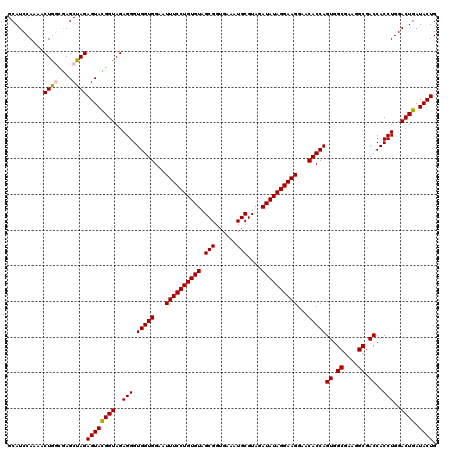
| Location | 722,761 – 722,881 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722761 120 + 6264404 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >pau.2632 1581136 120 - 6537648 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >pss.496 965352 120 + 6093698 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >ppk.94 172037 120 + 6181863 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >pel.69 116157 120 + 5888780 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >pfo.1282 2550195 120 + 6438405 GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). ( -36.10) >consensus GGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGA (...((((((((((.(((......)))....))))))))))...).(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))). (-36.10 = -36.10 + 0.00)
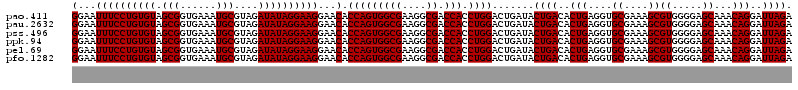
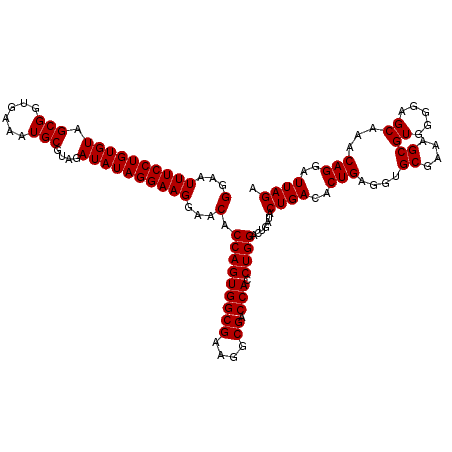
| Location | 722,761 – 722,881 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722761 120 - 6264404 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >pau.2632 1581136 120 + 6537648 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >pss.496 965352 120 - 6093698 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >ppk.94 172037 120 - 6181863 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >pel.69 116157 120 - 5888780 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >pfo.1282 2550195 120 - 6438405 UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... ( -25.40) >consensus UCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUCC .....((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))...... (-25.40 = -25.40 + 0.00)

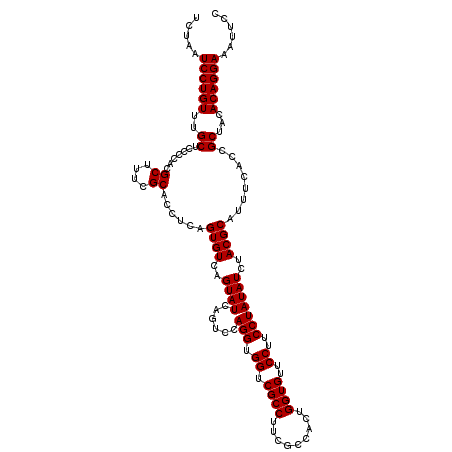
| Location | 722,841 – 722,961 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -37.91 |
| Energy contribution | -37.13 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722841 120 + 6264404 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUA ..........((....))(((...(((...((((.........)))).(((((.(((.((....)).))).)))))((((((((((((.....))))))))))))...)))..))).... ( -39.40) >pau.2632 1581216 120 - 6537648 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUA ..........((....))(((...(((...((((.........)))).(((((.(((.((....)).))).)))))((((((((((((.....))))))))))))...)))..))).... ( -39.40) >pss.496 965432 120 + 6093698 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUA .......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))). ( -40.10) >ppk.94 172117 120 + 6181863 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUA .......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))). ( -35.80) >pel.69 116237 120 + 5888780 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUA .......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))). ( -35.80) >pfo.1282 2550275 120 + 6438405 ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUA .......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))). ( -40.10) >consensus ACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUA .......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))). (-37.91 = -37.13 + -0.77)

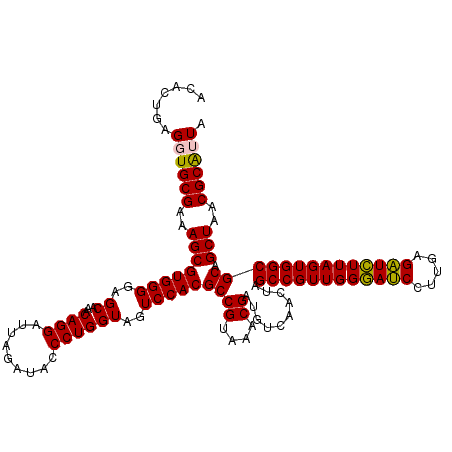
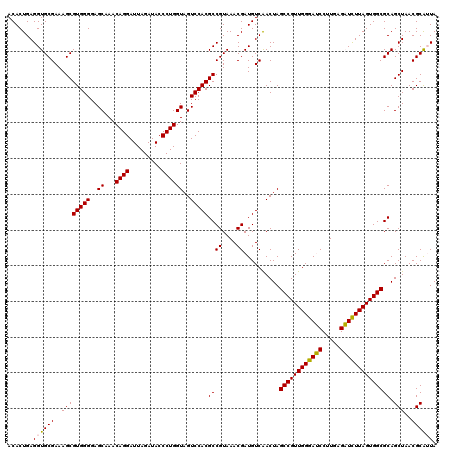
| Location | 722,841 – 722,961 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -36.27 |
| Energy contribution | -35.72 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722841 120 - 6264404 UAUCGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUCGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ....((((..((((.(((.....((((.....)))).....)))))))..)).........(((((((.....((((((.....))))))........)))))))...)).......... ( -34.12) >pau.2632 1581216 120 + 6537648 UAUCGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUCGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ....((((..((((.(((.....((((.....)))).....)))))))..)).........(((((((.....((((((.....))))))........)))))))...)).......... ( -34.12) >pss.496 965432 120 - 6093698 UAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ...(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))......... ( -38.92) >ppk.94 172117 120 - 6181863 UAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ...(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))......... ( -34.72) >pel.69 116237 120 - 5888780 UAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ...(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))......... ( -34.72) >pfo.1282 2550275 120 - 6438405 UAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ...(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))......... ( -38.92) >consensus UAAUGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGU ....((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)).......... (-36.27 = -35.72 + -0.55)
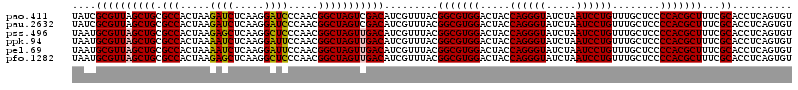
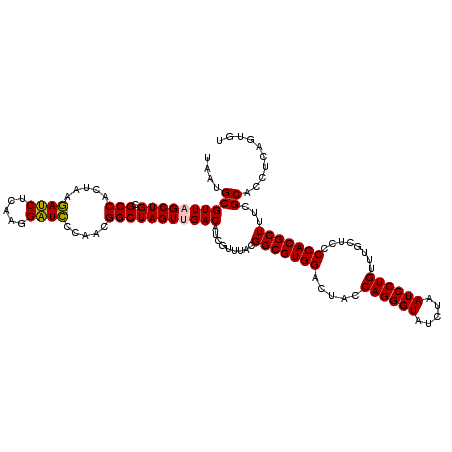
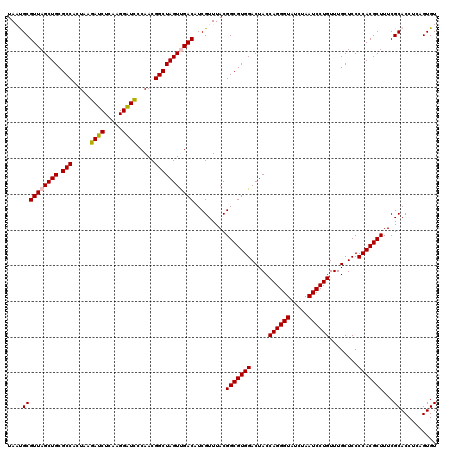
| Location | 722,881 – 723,001 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -44.43 |
| Energy contribution | -45.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722881 120 + 6264404 UACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -46.40) >pau.2632 1581256 120 - 6537648 UACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -46.40) >pss.496 965472 120 + 6093698 UACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -46.60) >ppk.94 172157 120 + 6181863 UACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -42.30) >pel.69 116277 120 + 5888780 UACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -42.30) >pfo.1282 2550315 120 + 6438405 UACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) ( -46.60) >consensus UACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUC ..(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))) (-44.43 = -45.10 + 0.67)
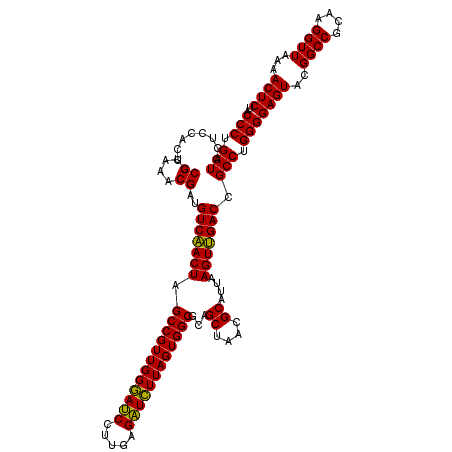
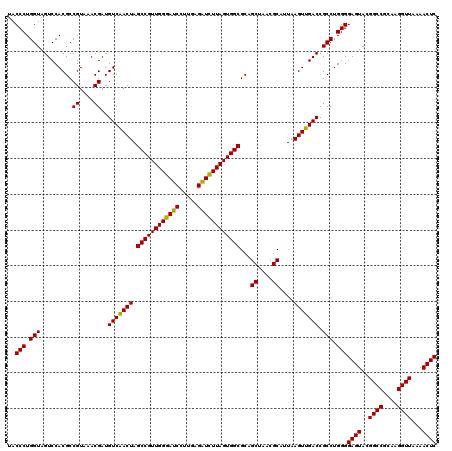
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:07:33 2007