
| Sequence ID | pao.411 |
|---|---|
| Location | 721,593 – 722,163 |
| Length | 570 |
| Max. P | 0.998970 |

| Location | 721,593 – 721,695 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.78 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -6.42 |
| Energy contribution | -6.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
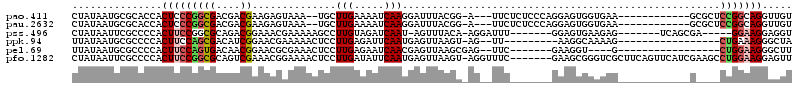
>pao.411 721593 102 + 6264404 CUAUAAUGCGCACCACUCCCGGCGACGACGAAGAGUAAA--UGCUUGAAAAUCAAGGAUUUACGG-A---UUCUCUCCCAGGAGUGGUGAA------------GCGCUCCGGCAGGUUGU ..(((((((.(((((((((.((.((.((.(((..(((((--(.((((.....)))).))))))..-.---))))))))).)))))))))..------------))((....))..))))) ( -40.40) >pau.2632 1579968 102 - 6537648 CUAUAAUGCGCACCACUCCCGGCGACGACGAAGAGUAAA--UGCUUGAAAAUCAAGGAUUUACGG-A---UUCUCUCCCAGGAGUGGUGAA------------GCGCUCCGGCAGGUUGU ..(((((((.(((((((((.((.((.((.(((..(((((--(.((((.....)))).))))))..-.---))))))))).)))))))))..------------))((....))..))))) ( -40.40) >pss.496 964202 99 + 6093698 CUAUAAUUCGCCCCACUUCCGGCGCAGACGGAAACGAAAAAGCCUUGUAGAUCAAU-AGUUUACA-AGGAUUU-------GGAGUGAAGAG-------UCAGCGA-----GGAAGGAGGU ...........((..(((((..(((.(((....((...(((.((((((((((....-.)))))))-))).)))-------...)).....)-------)).))).-----)))))..)). ( -33.30) >ppk.94 170911 91 + 6181863 UUAUAAUGCGCCCCACUUCCAGCGACAUCGGAACGAAAAACUCCUUGAGAUUCAAUGAGUUAAGU-AG--UU---------AAGGCAAAAG-----------------CUGAAAGGGCUA .........((((...((.((((....((..(((....(((((.(((.....))).)))))....-.)--))---------..)).....)-----------------))).)))))).. ( -20.80) >pel.69 115044 90 + 5888780 UUAUAAUGCGCCCCACUUCCAGUGACAACGGAACGCGAAACUCCUUGAGAAUCAACGAGUUAAGCGAG--UUC-------GAAGGU----G-----------------CUGGAAGGGCUU .........((((...(((((((..(..((((.(((..(((((.(((.....))).)))))..)))..--)))-------)..)..----)-----------------)))))))))).. ( -33.80) >pfo.1282 2549014 112 + 6438405 CUAUAAUUCGCCCCACUUCCGGCGCAGUCGAAACGGAAAACUCCUUGAUAUUCAAUGAGUUAAGU-AGGUUUC-------GAAGCGGGUCGCUUCAGUUCAUCGAAGCCUGGAAGGAGUU ...............((((((((((..(((((((..(.(((((.(((.....))).)))))...)-..)))))-------)).)))....(((((........))))))))))))..... ( -34.30) >consensus CUAUAAUGCGCCCCACUUCCGGCGACGACGAAAAGGAAAACUCCUUGAAAAUCAAUGAGUUAAGG_AG__UUC_______GAAGUGGAGAG____________G____CUGGAAGGACGU ...............(((((((((....))..............(((.....))).....................................................)))))))..... ( -6.42 = -6.78 + 0.36)
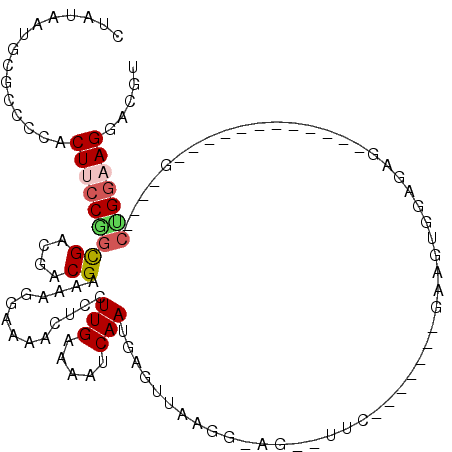
| Location | 721,593 – 721,695 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.78 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -6.49 |
| Energy contribution | -6.47 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721593 102 - 6264404 ACAACCUGCCGGAGCGC------------UUCACCACUCCUGGGAGAGAA---U-CCGUAAAUCCUUGAUUUUCAAGCA--UUUACUCUUCGUCGUCGCCGGGAGUGGUGCGCAUUAUAG ....((....)).((((------------...((((((((((((((((((---.-..((((((.((((.....)))).)--)))))..))).)).)).)))))))))))))))....... ( -40.60) >pau.2632 1579968 102 + 6537648 ACAACCUGCCGGAGCGC------------UUCACCACUCCUGGGAGAGAA---U-CCGUAAAUCCUUGAUUUUCAAGCA--UUUACUCUUCGUCGUCGCCGGGAGUGGUGCGCAUUAUAG ....((....)).((((------------...((((((((((((((((((---.-..((((((.((((.....)))).)--)))))..))).)).)).)))))))))))))))....... ( -40.60) >pss.496 964202 99 - 6093698 ACCUCCUUCC-----UCGCUGA-------CUCUUCACUCC-------AAAUCCU-UGUAAACU-AUUGAUCUACAAGGCUUUUUCGUUUCCGUCUGCGCCGGAAGUGGGGCGAAUUAUAG .(((((((((-----.(((.((-------(.....((...-------(((.(((-((((....-.......))))))).)))...))....))).)))..))))).)))).......... ( -26.20) >ppk.94 170911 91 - 6181863 UAGCCCUUUCAG-----------------CUUUUGCCUU---------AA--CU-ACUUAACUCAUUGAAUCUCAAGGAGUUUUUCGUUCCGAUGUCGCUGGAAGUGGGGCGCAUUAUAA ..((((((((((-----------------(.........---------..--..-....(((((.(((.....))).)))))..(((...)))....))))))...)))))......... ( -22.30) >pel.69 115044 90 - 5888780 AAGCCCUUCCAG-----------------C----ACCUUC-------GAA--CUCGCUUAACUCGUUGAUUCUCAAGGAGUUUCGCGUUCCGUUGUCACUGGAAGUGGGGCGCAUUAUAA ..((((((((((-----------------.----.....(-------(..--..(((..(((((.(((.....))).)))))..)))...))......)))))...)))))......... ( -30.80) >pfo.1282 2549014 112 - 6438405 AACUCCUUCCAGGCUUCGAUGAACUGAAGCGACCCGCUUC-------GAAACCU-ACUUAACUCAUUGAAUAUCAAGGAGUUUUCCGUUUCGACUGCGCCGGAAGUGGGGCGAAUUAUAG ..((((((((..((((((......))))))....(((.((-------(((((..-....(((((.(((.....))).)))))....)))))))..)))..))))).)))........... ( -36.10) >consensus AAAACCUUCCAG____C____________CUCACCACUCC_______GAA__CU_ACUUAACUCAUUGAUUUUCAAGGAGUUUUACGUUCCGUCGUCGCCGGAAGUGGGGCGCAUUAUAG ...............................((((((((..........................(((.....))).............(((.......))))))))))).......... ( -6.49 = -6.47 + -0.03)

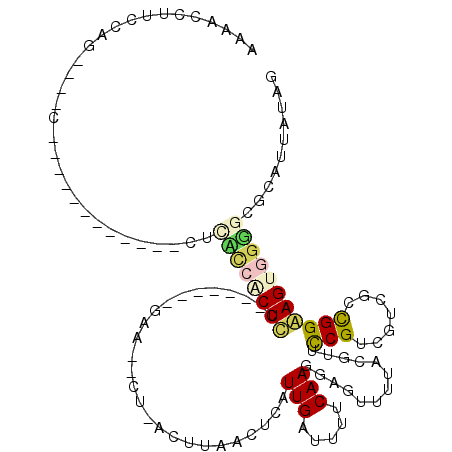
| Location | 721,667 – 721,746 |
|---|---|
| Length | 79 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 37.65 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -2.30 |
| Energy contribution | -3.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.07 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575358 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.411 721667 79 + 6264404 GGAGUGGUGAA------------GCGCUCCGGCAGGUUGUU--------------------CGCCGCAGCGGUUCGGU-------CCCGAAAGGGGCUCGAAACGAAGCUUCAUCGAG ....(((((((------------(((((.((((.((....)--------------------))))).)))(.((((((-------(((....))))).)))).)...))))))))).. ( -40.40) >pau.2632 1580042 79 - 6537648 GGAGUGGUGAA------------GCGCUCCGGCAGGUUGUU--------------------CGCCGCAGCGGUUCGGU-------CCCGAAAGGGGCUCGAAACGAAGCUUCAUCGAG ....(((((((------------(((((.((((.((....)--------------------))))).)))(.((((((-------(((....))))).)))).)...))))))))).. ( -40.40) >ppk.94 170980 51 + 6181863 -AAGGCAAAAG-----------------CUGAAAGGGCUAC-----------------------------GAUCGAAU------GAUCGAAAGCGGUUGAGA--------------UG -...((...((-----------------((.....)))).(-----------------------------((((....------)))))...))........--------------.. ( -10.60) >pfo.1282 2549086 104 + 6438405 GAAGCGGGUCGCUUCAGUUCAUCGAAGCCUGGAAGGAGUUGGAAGUUCCGGUUUUUUGGGUGCUUUCAACGGUUCGAUCUUCUCGGUCGAAAGCGGAGAAAA--------------GA ((((((...)))))).....((((((.((.....)).((((((((((((((....))))).)))))))))..)))))).(((((.(.(....)).)))))..--------------.. ( -36.00) >consensus GAAGUGGUGAA____________GCGCUCCGGAAGGUUGUU____________________CGCCGCAGCGGUUCGAU_______CCCGAAAGCGGCUCAAA______________AG .....................................................................................(((....)))....................... ( -2.30 = -3.30 + 1.00)

| Location | 721,667 – 721,746 |
|---|---|
| Length | 79 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 37.65 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -2.92 |
| Energy contribution | -2.30 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.12 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659404 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.411 721667 79 - 6264404 CUCGAUGAAGCUUCGUUUCGAGCCCCUUUCGGG-------ACCGAACCGCUGCGGCG--------------------AACAACCUGCCGGAGCGC------------UUCACCACUCC ...(.((((((...(.((((..(((.....)))-------..)))).)(((.(((((--------------------.......))))).)))))------------)))).)..... ( -31.00) >pau.2632 1580042 79 + 6537648 CUCGAUGAAGCUUCGUUUCGAGCCCCUUUCGGG-------ACCGAACCGCUGCGGCG--------------------AACAACCUGCCGGAGCGC------------UUCACCACUCC ...(.((((((...(.((((..(((.....)))-------..)))).)(((.(((((--------------------.......))))).)))))------------)))).)..... ( -31.00) >ppk.94 170980 51 - 6181863 CA--------------UCUCAACCGCUUUCGAUC------AUUCGAUC-----------------------------GUAGCCCUUUCAG-----------------CUUUUGCCUU- ..--------------........(((..(((((------....))))-----------------------------).))).......(-----------------(....))...- ( -9.60) >pfo.1282 2549086 104 - 6438405 UC--------------UUUUCUCCGCUUUCGACCGAGAAGAUCGAACCGUUGAAAGCACCCAAAAAACCGGAACUUCCAACUCCUUCCAGGCUUCGAUGAACUGAAGCGACCCGCUUC ..--------------........((((((((((((.....)))....)))))))))............((((...........))))..((((((......)))))).......... ( -22.50) >consensus CU______________UUCGAGCCCCUUUCGAG_______ACCGAACCGCUGCGGCG____________________AAAAACCUGCCAGAGCGC____________CUCACCACUCC ...........................(((((.........)))))........................................................................ ( -2.92 = -2.30 + -0.62)


| Location | 721,746 – 721,849 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.06 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.86 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721746 103 + 6264404 GUGCUUGACAGCGAAUUUGAACGCUGUAGAAUGCGCCUCCCGCUGAUCGGAAGAUGGUUUGAAGGUC--GGCGCAAGCGGUUGAGUAGA--AAAGA-----------AAAUUUUC--GAA .((((..((((((........))))...(..((((((..(((((.(((....)))))).....))..--))))))..).))..))))((--(((..-----------...)))))--... ( -34.00) >pau.2632 1580121 103 - 6537648 GUGCUUGACAGCGAAUUUGAACGCUGUAGAAUGCGCCUCCCGCUGAUCGGAAGAUGGUUUGAAGGUC--GGCGCAAGCGGUUGAGUAGA--AAAGA-----------AAAUUUUC--GAA .((((..((((((........))))...(..((((((..(((((.(((....)))))).....))..--))))))..).))..))))((--(((..-----------...)))))--... ( -34.00) >pss.496 964335 116 + 6093698 GAGUUUGACACGCGGUUGUAACGCUGUAGAAUUCGCCUCCCGCUGAUGAGCAACUCAAGAGUUGAUCGCAGCGCAAGUGGUUGAAGUUG-CAAAGGAAACUUUG---AAAACUUCUUGAA ((((((.(((.(((.......)))))).))))))(((.(.(((((.(((.(((((....))))).))))))))...).))).((((((.-(((((....)))))---..))))))..... ( -42.10) >ppk.94 171031 106 + 6181863 AUGGUUGACAGCGCUUCCAAGUGCUGUAUGAUUCGCAUCCCGCU-ACGAGAGAUCGCAGCG--------AGC-CAAGUGUUUGAAGCUA--AACGAGUUUCUCGCAAAAAACUUC--AAA .(((((.((((((((....)))))))).............((((-.(((....))).))))--------)))-)).(((...((((((.--....)))))).)))..........--... ( -32.70) >pel.69 115162 95 + 5888780 GUGGUUGACAGCAAGUUGUAACGCUGUAUGAUUCGCCUCCCGCU-ACGAGAGAUCGCAGCG--------AGU-UAAGCGGUUGAAGUUG--AACGA-----------AAAACUUC--GAA .((.(((((.((.(((..((......))..))).))....((((-.(((....))).))))--------.))-))).)).((((((((.--.....-----------..))))))--)). ( -25.60) >pfo.1282 2549190 106 + 6438405 GGUGUUGACAGCAGCGUGUAACGCUGUAGAAUUCGCCUCCCGCUAACGAGAGAUCGGA-----------AGCGCAAGUGGUUGAAGUUGUUGAAGAAAUCUUCG---AAAACUUCUGAAA ((((....(.((((((.....)))))).)....))))...((((..(((....)))..-----------)))).........((((((.((((((....)))))---).))))))..... ( -35.90) >consensus GUGGUUGACAGCGAAUUGUAACGCUGUAGAAUUCGCCUCCCGCUGACGAGAAAUCGGAGCG________AGCGCAAGCGGUUGAAGUUA__AAAGA___________AAAACUUC__GAA ((((...(((((..........))))).....))))...(((((...............................)))))........................................ ( -8.11 = -8.86 + 0.75)

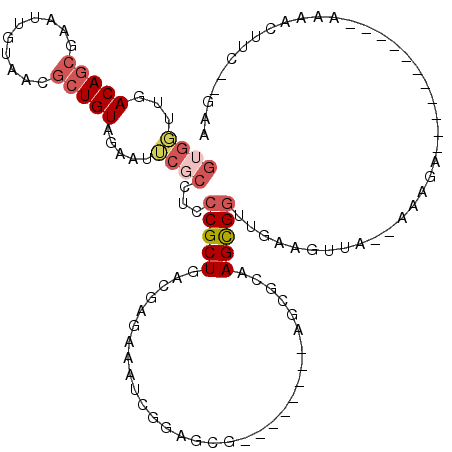
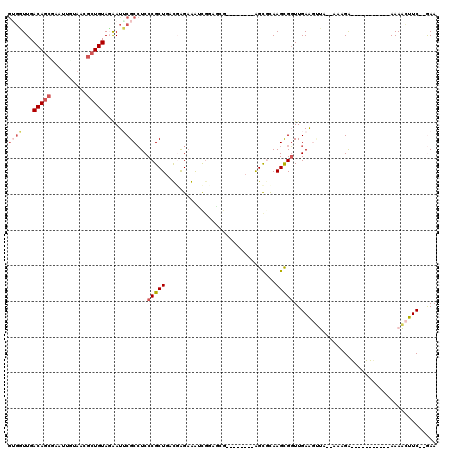
| Location | 721,746 – 721,849 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.06 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -9.00 |
| Energy contribution | -10.08 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.29 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721746 103 - 6264404 UUC--GAAAAUUU-----------UCUUU--UCUACUCAACCGCUUGCGCC--GACCUUCAAACCAUCUUCCGAUCAGCGGGAGGCGCAUUCUACAGCGUUCAAAUUCGCUGUCAAGCAC ...--(((((...-----------..)))--)).........(((((((((--(.....)........(((((.....)))))))))).....((((((........)))))).)))).. ( -28.10) >pau.2632 1580121 103 + 6537648 UUC--GAAAAUUU-----------UCUUU--UCUACUCAACCGCUUGCGCC--GACCUUCAAACCAUCUUCCGAUCAGCGGGAGGCGCAUUCUACAGCGUUCAAAUUCGCUGUCAAGCAC ...--(((((...-----------..)))--)).........(((((((((--(.....)........(((((.....)))))))))).....((((((........)))))).)))).. ( -28.10) >pss.496 964335 116 - 6093698 UUCAAGAAGUUUU---CAAAGUUUCCUUUG-CAACUUCAACCACUUGCGCUGCGAUCAACUCUUGAGUUGCUCAUCAGCGGGAGGCGAAUUCUACAGCGUUACAACCGCGUGUCAAACUC (((..((((((..---(((((....)))))-.)))))).....(((.(((((.((.(((((....))))).))..))))).)))..)))....((((((.......))).)))....... ( -31.10) >ppk.94 171031 106 - 6181863 UUU--GAAGUUUUUUGCGAGAAACUCGUU--UAGCUUCAAACACUUG-GCU--------CGCUGCGAUCUCUCGU-AGCGGGAUGCGAAUCAUACAGCACUUGGAAGCGCUGUCAACCAU (((--((((((....(((((...))))).--.)))))))))....((-(((--------((((((((....))))-))))))...........(((((.((....)).)))))...))). ( -37.70) >pel.69 115162 95 - 5888780 UUC--GAAGUUUU-----------UCGUU--CAACUUCAACCGCUUA-ACU--------CGCUGCGAUCUCUCGU-AGCGGGAGGCGAAUCAUACAGCGUUACAACUUGCUGUCAACCAC ...--((((((..-----------.....--.))))))...(((((.-.((--------((((((((....))))-)))))))))))......((((((........))))))....... ( -32.60) >pfo.1282 2549190 106 - 6438405 UUUCAGAAGUUUU---CGAAGAUUUCUUCAACAACUUCAACCACUUGCGCU-----------UCCGAUCUCUCGUUAGCGGGAGGCGAAUUCUACAGCGUUACACGCUGCUGUCAACACC .(((.((((((..---.((((....))))...)))))).....(((.((((-----------..(((....)))..)))).)))..))).....(((((.....)))))........... ( -31.40) >consensus UUC__GAAGUUUU___________UCUUU__CAACUUCAACCACUUGCGCU________CACUCCAAUCUCUCAUCAGCGGGAGGCGAAUUCUACAGCGUUACAACUCGCUGUCAACCAC .....((((((.....................)))))).............................((((((......))))))........((((((........))))))....... ( -9.00 = -10.08 + 1.09)

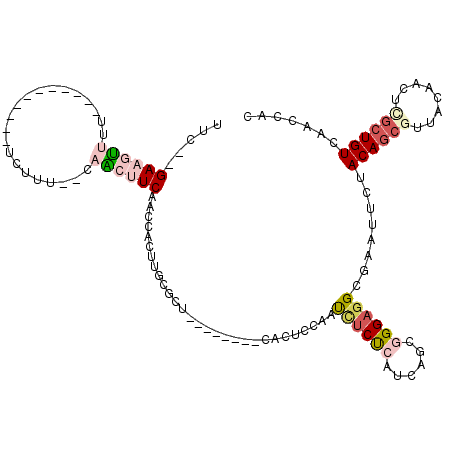
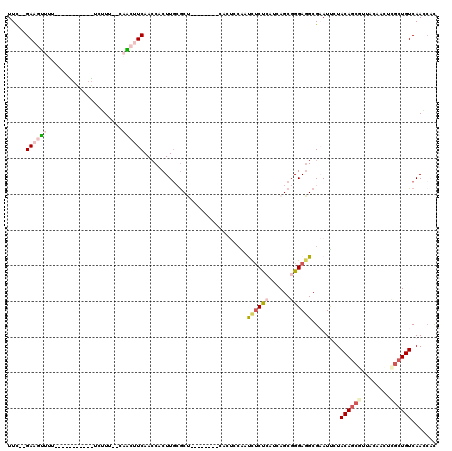
| Location | 721,786 – 721,889 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.36 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.45 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.26 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721786 103 + 6264404 CGCUGAUCGGAAGAUGGUUUGAAGGUC--GGCGCAAGCGGUUGAGUAGA--AAAGA-----------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGC ((((((((....((....))...))))--))))((((((........((--(((..-----------...)))))--........))))))((((..(....)....))))......... ( -26.69) >pau.2632 1580161 103 - 6537648 CGCUGAUCGGAAGAUGGUUUGAAGGUC--GGCGCAAGCGGUUGAGUAGA--AAAGA-----------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGC ((((((((....((....))...))))--))))((((((........((--(((..-----------...)))))--........))))))((((..(....)....))))......... ( -26.69) >pss.496 964375 116 + 6093698 CGCUGAUGAGCAACUCAAGAGUUGAUCGCAGCGCAAGUGGUUGAAGUUG-CAAAGGAAACUUUG---AAAACUUCUUGAAAUAACCGCUUGACAGAUACAGAGGACGCUGUAGAAUGCGC (((((.(((.(((((....))))).))))))))(((((((((((((((.-(((((....)))))---..)))))).......))))))))).((..(((((......)))))...))... ( -43.21) >ppk.94 171071 106 + 6181863 CGCU-ACGAGAGAUCGCAGCG--------AGC-CAAGUGUUUGAAGCUA--AACGAGUUUCUCGCAAAAAACUUC--AAAAUAAACGCUUGACAGGCUCUGAGGAAAGCGUAGAAUGCGC .(((-.(((....))).)))(--------(((-(((((((((((((...--..((((...)))).......))))--).....)))))))....)))))........((((.....)))) ( -28.70) >pel.69 115202 95 + 5888780 CGCU-ACGAGAGAUCGCAGCG--------AGU-UAAGCGGUUGAAGUUG--AACGA-----------AAAACUUC--GAAAAUAACGCUUGACACGAAAUGAGGUUAGCGUAGAAUGCGC ..((-(((...((((.((.((--------.((-((((((.((((((((.--.....-----------..))))))--))......)))))))).))...)).))))..)))))....... ( -31.70) >pfo.1282 2549230 106 + 6438405 CGCUAACGAGAGAUCGGA-----------AGCGCAAGUGGUUGAAGUUGUUGAAGAAAUCUUCG---AAAACUUCUGAAAAUAAUCACUUGACAGCAAAUGAGGCUGCUGUAGAAUGCGC ((((..(((....)))..-----------))))(((((((((((((((.((((((....)))))---).)))))).......)))))))))((((((........))))))......... ( -36.71) >consensus CGCUGACGAGAAAUCGGAGCG________AGCGCAAGCGGUUGAAGUUA__AAAGA___________AAAACUUC__GAAAAAAACGCUUGACAGAAAAAGAGGUUGCUGUAGAAUGCGC (((..............................((((((...((((((.....................))))))..........))))))((((............)))).....))). ( -8.53 = -8.45 + -0.08)

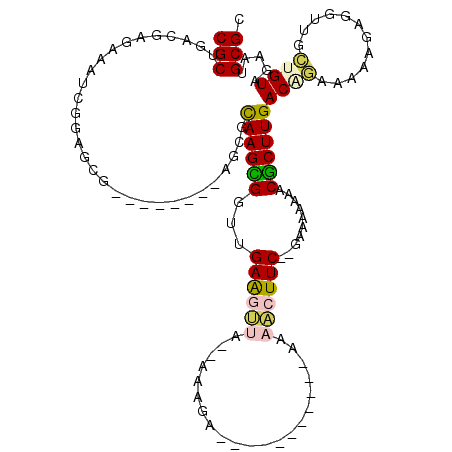
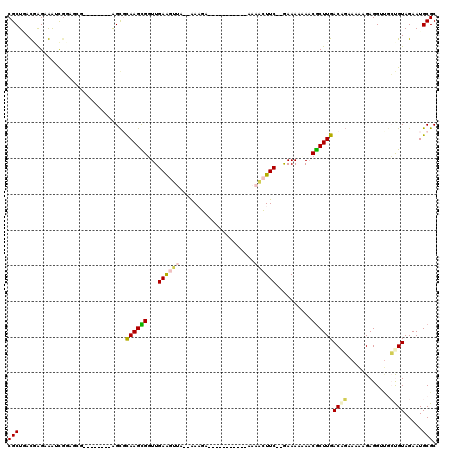
| Location | 721,824 – 721,927 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721824 103 + 6264404 UUGAGUAGA--AAAGA-----------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACGAAA--GCCUUGACCAACUGCUCUU ..((((((.--.....-----------....((((--(..((......))..)))))...((((((((.((.....))))))))))(((..((.(....--).))..)))..)))))).. ( -34.10) >pau.2632 1580199 103 - 6537648 UUGAGUAGA--AAAGA-----------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACGAAA--GCCUUGACCAACUGCUCUU ..((((((.--.....-----------....((((--(..((......))..)))))...((((((((.((.....))))))))))(((..((.(....--).))..)))..)))))).. ( -34.10) >pss.496 964415 114 + 6093698 UUGAAGUUG-CAAAGGAAACUUUG---AAAACUUCUUGAAAUAACCGCUUGACAGAUACAGAGGACGCUGUAGAAUGCGC-GCCUCGGUUGAGACGAAA-GGUCUUAACCCACCGCUCUU ..((((((.-(((((....)))))---..))))))...........((............((((.(((.((.....))))-)))))(((((((((....-.)))))))))....)).... ( -38.40) >ppk.94 171101 115 + 6181863 UUGAAGCUA--AACGAGUUUCUCGCAAAAAACUUC--AAAAUAAACGCUUGACAGGCUCUGAGGAAAGCGUAGAAUGCGC-GCCUCGGUUGAGACGAAAAGCUCUUAACCAAACGCUCUU ..((((((.--....))))))..((..........--.........(((.....)))...((((...((((.....))))-.))))(((((((((.....).))))))))....)).... ( -31.00) >pel.69 115232 104 + 5888780 UUGAAGUUG--AACGA-----------AAAACUUC--GAAAAUAACGCUUGACACGAAAUGAGGUUAGCGUAGAAUGCGC-GCCUCGGUUGAGACGAAACGCUCUUAACCAAUCGCUCUU ((((((((.--.....-----------..))))))--)).......((............(((((..((((.....))))-)))))((((((((((...)).))))))))....)).... ( -31.30) >pfo.1282 2549259 115 + 6438405 UUGAAGUUGUUGAAGAAAUCUUCG---AAAACUUCUGAAAAUAAUCACUUGACAGCAAAUGAGGCUGCUGUAGAAUGCGC-GCCUCGGUUGAGACGAAA-GAUCUUAACCAACCGCUCUU ..((((((.((((((....)))))---).))))))..................(((....((((((((........))).-)))))(((((((((....-).))))))))....)))... ( -41.00) >consensus UUGAAGUUA__AAAGA___________AAAACUUC__GAAAAAAACGCUUGACAGAAAAAGAGGUUGCUGUAGAAUGCGC_GCCUCGGUUGAGACGAAA_GGUCUUAACCAACCGCUCUU ..............................................((............(((((...(((.....)))..)))))((((((((........))))))))....)).... (-16.43 = -16.73 + 0.31)

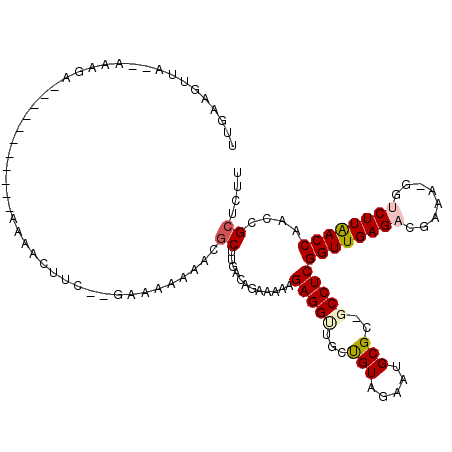
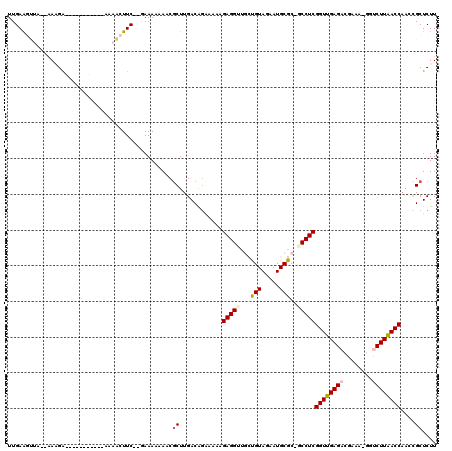
| Location | 721,889 – 722,006 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.66 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -30.39 |
| Energy contribution | -29.75 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721889 117 + 6264404 GGCCUCGGUUGAGACGAAA--GCCUUGACCAACUGCUCUUUAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUA .(((..(((..((.(....--).))..))).............((...(((((......)))))..)))))(((((((.((.....(((((((((.-...))))))))))).))))))). ( -40.40) >pau.2632 1580264 117 - 6537648 GGCCUCGGUUGAGACGAAA--GCCUUGACCAACUGCUCUUUAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUA .(((..(((..((.(....--).))..))).............((...(((((......)))))..)))))(((((((.((.....(((((((((.-...))))))))))).))))))). ( -40.40) >pss.496 964491 115 + 6093698 -GCCUCGGUUGAGACGAAA-GGUCUUAACCCACCGCUCUUUAAC-AACUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGUGUCAGACUGA-AGUCAAC-AGAUUAUCAGCAACCCAAGUU -((...(((((((((....-.)))))))))....))........-((((((((......))))...((((((((.(((((.(..(((...-.)))...-).))))).))).))))))))) ( -31.00) >ppk.94 171177 119 + 6181863 -GCCUCGGUUGAGACGAAAAGCUCUUAACCAAACGCUCUUUAACAAAUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUAUGGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUG -(((..(((((((((.....).)))))))).............((...(((((......)))))..)))))((((((((.....(.(((((((((.....)))))))))).)))))))). ( -38.20) >pel.69 115297 119 + 5888780 -GCCUCGGUUGAGACGAAACGCUCUUAACCAAUCGCUCUUUAACAAAUUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUACGGGCUGAUAGUCACCAAGAUUAUCAGCAUCACAAGUG -.....((((((((((...)).))))))))....((((....((.(((((.......))))).))..))))((((((((......((((((((((.....)))))))))).)))))))). ( -40.70) >pfo.1282 2549336 117 + 6438405 -GCCUCGGUUGAGACGAAA-GAUCUUAACCAACCGCUCUUUAAC-AACUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGAGUCAGACUGAUAGUCAACAAGAUUAUCAGCAUCACAAGUU -((((.(((((((((....-).))))))))............((-(...((((......)))).)))))))(((((((..(.....(((((((((.....))))))))))..))))))). ( -35.70) >consensus _GCCUCGGUUGAGACGAAA_GGUCUUAACCAACCGCUCUUUAACAAAUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUCAGACUGAUAGUCAACAAGAUUAUCAGCAACACAAGUA .((((.((((((((........))))))))..................(((((......)))))...))))(((((((......(.(((((((((.....))))))))))..))))))). (-30.39 = -29.75 + -0.64)
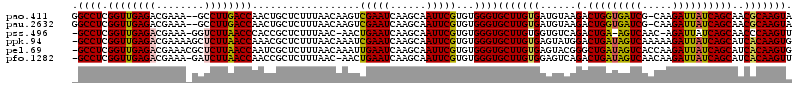
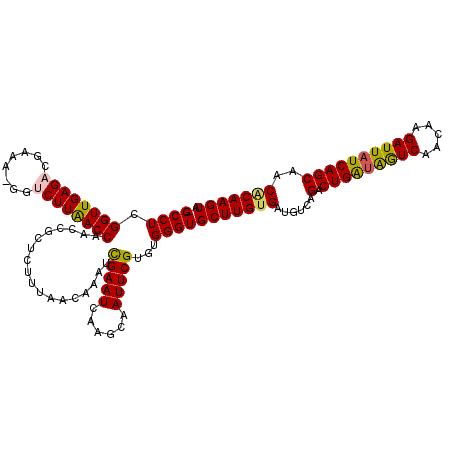
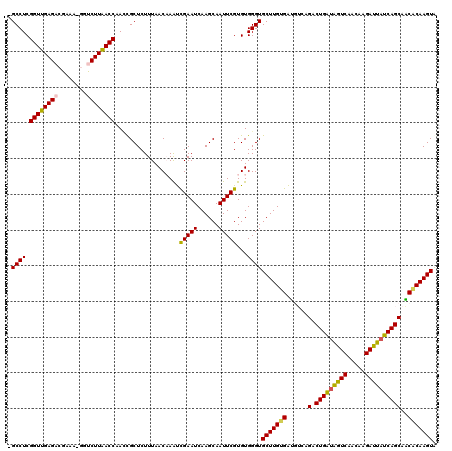
| Location | 721,927 – 722,045 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -25.36 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721927 118 + 6264404 UAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUAAC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGA .......((((((((.......((.(((..((((((((.((.....(((((((((.-...))))))))))).)))))))).)-)).)))).))))))........(((((.....))))) ( -36.61) >pau.2632 1580302 118 - 6537648 UAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUAAC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGA .......((((((((.......((.(((..((((((((.((.....(((((((((.-...))))))))))).)))))))).)-)).)))).))))))........(((((.....))))) ( -36.61) >pss.496 964529 104 + 6093698 UAAC-AACUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGUGUCAGACUGA-AGUCAAC-AGAUUAUCAGCAACCCAAGUUACUCCGCGAGAAAUCAAAGAUGUAACC------------- ..((-(.((((((......))))(((.(((((((((.(((....(.((((-((((...-.)))).))))).))))))).))))))))((....))..)).)))....------------- ( -25.80) >ppk.94 171216 106 + 6181863 UAACAAAUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUAUGGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUGACCAUGCGAGAAAUCACAUA-GUCAUU------------- ....................(((((((((..((((((((.....(.(((((((((.....)))))))))).))))))))..)))))))))..........-......------------- ( -35.30) >pel.69 115336 106 + 5888780 UAACAAAUUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUACGGGCUGAUAGUCACCAAGAUUAUCAGCAUCACAAGUGGCCAUGCGAGAAAUCACAUA-GUCAUU------------- ....................(((((((((.(((((((((......((((((((((.....)))))))))).))))))))).)))))))))..........-......------------- ( -39.50) >pfo.1282 2549374 106 + 6438405 UAAC-AACUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGAGUCAGACUGAUAGUCAACAAGAUUAUCAGCAUCACAAGUUACUCCGCGAGAAAUCAAAGAUGUAACC------------- ..((-(.((((((......))))(((.(((((((((((..(.....(((((((((.....))))))))))..)))))).))))))))((....))..)).)))....------------- ( -30.20) >consensus UAACAAAUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUCAGACUGAUAGUCAACAAGAUUAUCAGCAACACAAGUAAC_ACGCGAGAAAUCAAAAAUGUAAUC_____________ ....................(((((((((.((((((((......(.(((((((((.....))))))))))..)))))))).))))))))).............................. (-25.36 = -26.08 + 0.73)


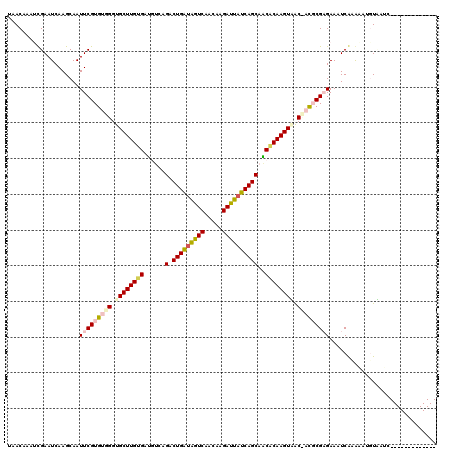
| Location | 721,927 – 722,045 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.91 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -16.87 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721927 118 - 6264404 UCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GUUACUUGCGUUGCUGAUAAUCUUG-CGAUCACCAGUCUUACAUCACAAGCACCCACACGAAUUGCUUGAUUCGACUUGUUA (((((.....)))))...(((..((((((....((((-(..(((.(.(((((..........)-))).).).)))..))).)).((((((...........))))))))))))...))). ( -23.60) >pau.2632 1580302 118 + 6537648 UCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GUUACUUGCGUUGCUGAUAAUCUUG-CGAUCACCAGUCUUACAUCACAAGCACCCACACGAAUUGCUUGAUUCGACUUGUUA (((((.....)))))...(((..((((((....((((-(..(((.(.(((((..........)-))).).).)))..))).)).((((((...........))))))))))))...))). ( -23.60) >pss.496 964529 104 - 6093698 -------------GGUUACAUCUUUGAUUUCUCGCGGAGUAACUUGGGUUGCUGAUAAUCU-GUUGACU-UCAGUCUGACACCACAAGCACCCACACGAAUUGCUUGAUUCAGUU-GUUA -------------((((((.(((.(((....))).)))))))))(((((.(((.......(-((((((.-...))).)))).....))))))))...(((((....)))))....-.... ( -25.50) >ppk.94 171216 106 - 6181863 -------------AAUGAC-UAUGUGAUUUCUCGCAUGGUCACUUGUGAUGCUGAUAAUCUUUUUGACUAUCAGUCCAUACUCACAAGCACCCACACGAAUUGCUUGAUUCGAUUUGUUA -------------..((((-(((((((....)))))))))))(((((((.(((((((.((.....)).)))))))......)))))))........((((((....))))))........ ( -34.30) >pel.69 115336 106 - 5888780 -------------AAUGAC-UAUGUGAUUUCUCGCAUGGCCACUUGUGAUGCUGAUAAUCUUGGUGACUAUCAGCCCGUACUCACAAGCACCCACACGAAUUGCUUGAUUCAAUUUGUUA -------------..((.(-(((((((....)))))))).))(((((((.(((((((.((.....)).)))))))......))))))).......((((((((.......)))))))).. ( -30.50) >pfo.1282 2549374 106 - 6438405 -------------GGUUACAUCUUUGAUUUCUCGCGGAGUAACUUGUGAUGCUGAUAAUCUUGUUGACUAUCAGUCUGACUCCACAAGCACCCACACGAAUUGCUUGAUUCAGUU-GUUA -------------((((((.(((.(((....))).))))))))).((.(.(((((((.((.....)).))))))).).))....((((((...........))))))........-.... ( -24.40) >consensus _____________GAUAACAUAUUUGAUUUCUCGCGU_GUAACUUGUGAUGCUGAUAAUCUUGUUGACUAUCAGUCUGACAUCACAAGCACCCACACGAAUUGCUUGAUUCAAUUUGUUA ...............((((.((((((......))))))))))........(((((((.((.....)).))))))).........((((((...........))))))............. (-16.87 = -17.18 + 0.31)

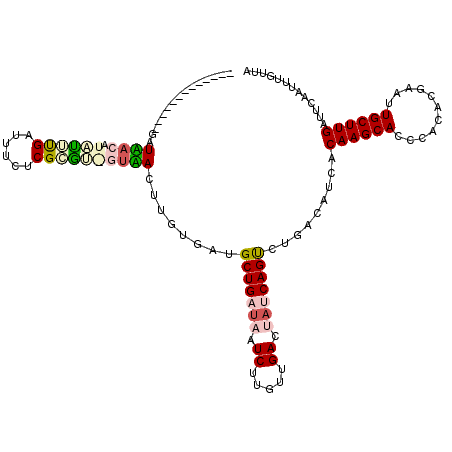
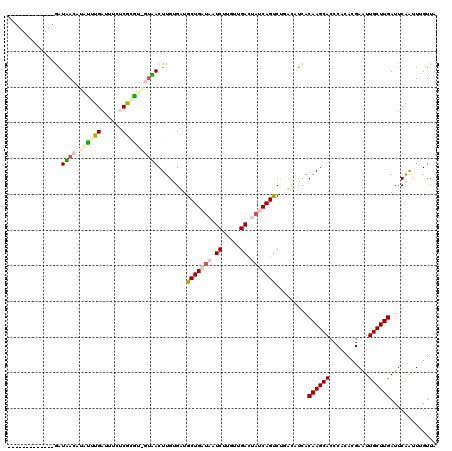
| Location | 721,967 – 722,085 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.31 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721967 118 + 6264404 GUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUAAC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCC ...((((((((((((.-...)))).(((((((((((...(((-.((((......)))).)))...(((((.....))))).)))).)))))))))).))))).((((((.....)))))) ( -42.70) >pau.2632 1580342 118 - 6537648 GUAAGACUGGUGAUCG-CAAGAUUAUCAGCAACGCAAGUAAC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCC ...((((((((((((.-...)))).(((((((((((...(((-.((((......)))).)))...(((((.....))))).)))).)))))))))).))))).((((((.....)))))) ( -42.70) >pss.496 964568 104 + 6093698 GUCAGACUGA-AGUCAAC-AGAUUAUCAGCAACCCAAGUUACUCCGCGAGAAAUCAAAGAUGUAACC--------------AACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCC ...(((((..-((((...-.)))).(((((((.(...(((((((...((....))...)).))))).--------------...).)))))))....))))).((((((.....)))))) ( -23.90) >ppk.94 171256 105 + 6181863 UAUGGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUGACCAUGCGAGAAAUCACAUA-GUCAUU--------------UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCC ..(((.(((((((((.....)))))))((((((.((((((((.(((.((....)).))).-))))))--------------)).)).)))).)))))......((((((.....)))))) ( -37.40) >pel.69 115376 105 + 5888780 UACGGGCUGAUAGUCACCAAGAUUAUCAGCAUCACAAGUGGCCAUGCGAGAAAUCACAUA-GUCAUU--------------UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCC ....(((((((((((.....)))))))((((((.((((((((.(((.((....)).))).-))))))--------------)).)).)))).)))).......((((((.....)))))) ( -39.60) >pfo.1282 2549413 106 + 6438405 GUCAGACUGAUAGUCAACAAGAUUAUCAGCAUCACAAGUUACUCCGCGAGAAAUCAAAGAUGUAACC--------------AACGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCC .((((.(((((((((.....))))))))).(((....(((((((...((....))...)).))))).--------------...)))..))))..........((((((.....)))))) ( -25.70) >consensus GUCAGACUGAUAGUCAACAAGAUUAUCAGCAACACAAGUAAC_ACGCGAGAAAUCAAAAAUGUAAUC______________UGCGAUUGCUGAGCCAAGUUUAGGGUUUCCUAAAAACCC ....(.(((((((((.....)))))))))).......(((((..((.(......).))...))))).......................((((((...))))))(((((.....))))). (-18.08 = -17.12 + -0.97)

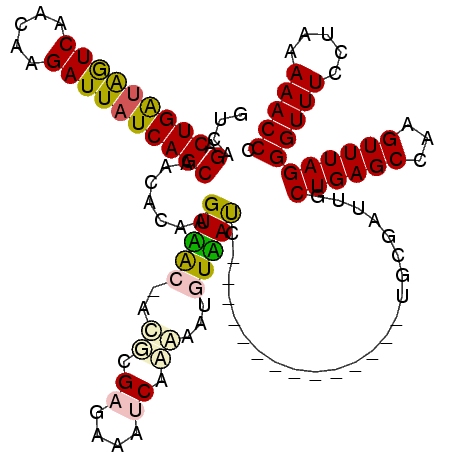
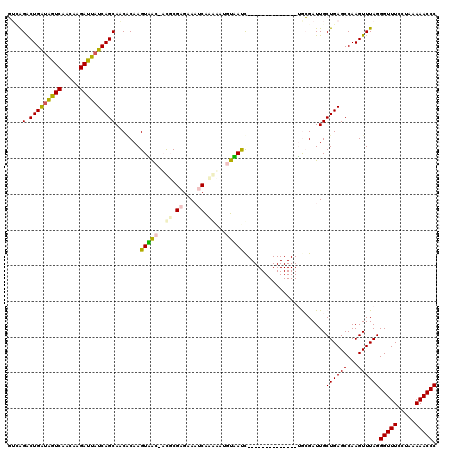
| Location | 721,967 – 722,085 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -19.36 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 721967 118 - 6264404 GGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GUUACUUGCGUUGCUGAUAAUCUUG-CGAUCACCAGUCUUAC ((((((.....)))))).....((((.(((((((.((((.(((((.....)))))...(((.((((......)))).-)))...)))))))))))..(((...-.)))..))))...... ( -33.90) >pau.2632 1580342 118 + 6537648 GGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GUUACUUGCGUUGCUGAUAAUCUUG-CGAUCACCAGUCUUAC ((((((.....)))))).....((((.(((((((.((((.(((((.....)))))...(((.((((......)))).-)))...)))))))))))..(((...-.)))..))))...... ( -33.90) >pss.496 964568 104 - 6093698 GGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGUU--------------GGUUACAUCUUUGAUUUCUCGCGGAGUAACUUGGGUUGCUGAUAAUCU-GUUGACU-UCAGUCUGAC ((((((.....))))))..(((.....(((((((((...--------------((((((.(((.(((....))).)))))))))..)))))))))......-)))(((.-...))).... ( -30.80) >ppk.94 171256 105 - 6181863 GGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA--------------AAUGAC-UAUGUGAUUUCUCGCAUGGUCACUUGUGAUGCUGAUAAUCUUUUUGACUAUCAGUCCAUA (((((((...)))))))......(((((.((((.((.((--------------(.((((-(((((((....))))))))))).))).))))))((((.((.....)).)))))).))).. ( -36.60) >pel.69 115376 105 - 5888780 GGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA--------------AAUGAC-UAUGUGAUUUCUCGCAUGGCCACUUGUGAUGCUGAUAAUCUUGGUGACUAUCAGCCCGUA (((((((...))))))).......(((((((((.((.((--------------(.((.(-(((((((....)))))))).)).))).)))))))).......(....).....))).... ( -34.80) >pfo.1282 2549413 106 - 6438405 GGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCGUU--------------GGUUACAUCUUUGAUUUCUCGCGGAGUAACUUGUGAUGCUGAUAAUCUUGUUGACUAUCAGUCUGAC (((((((...)))))))..(((..((.((((((.(((..--------------((((((.(((.(((....))).)))))))))..)))))))))...))..)))(((.....))).... ( -30.40) >consensus GGGUUUUGAAAAAACCCUAAACUUGGCUCAGCAAUCGCA______________GAUAACAUAUUUGAUUUCUCGCGU_GUAACUUGUGAUGCUGAUAAUCUUGUUGACUAUCAGUCUGAC (((((((...))))))).......(((((((((......................((((.((((((......))))))))))(....).))))))...((.....))......))).... (-19.36 = -18.58 + -0.77)
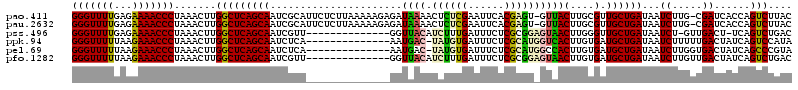
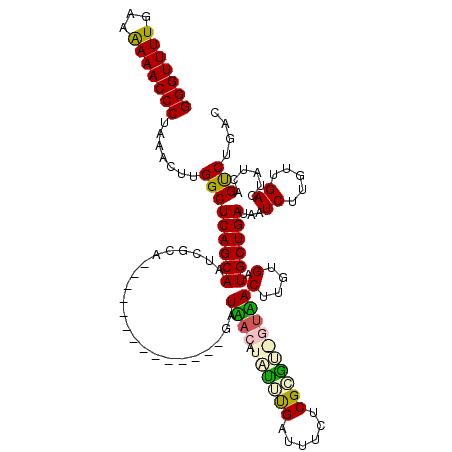
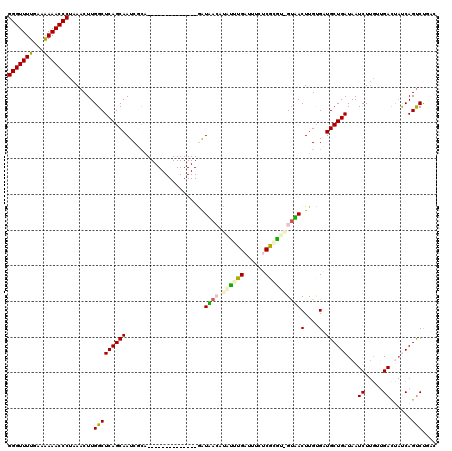
| Location | 722,006 – 722,123 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722006 117 + 6264404 AC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGA ..-.((((......)))).((....(((((.....)))))..)).....((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).)))))))). ( -38.10) >pau.2632 1580381 117 - 6537648 AC-ACUCGUGAAUUCGAGAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGA ..-.((((......)))).((....(((((.....)))))..)).....((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).)))))))). ( -38.10) >pss.496 964606 106 + 6093698 ACUCCGCGAGAAAUCAAAGAUGUAACC--------------AACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAGUUAGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGA .......((....))............--------------........(((((((.......((((((.....))))))......(((.(..((((....))))..).)))))))))). ( -28.90) >ppk.94 171296 103 + 6181863 ACCAUGCGAGAAAUCACAUA-GUCAUU--------------UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGA ...(((.((....)).)))(-(((...--------------...)))).((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).)))))))). ( -33.90) >pel.69 115416 103 + 5888780 GCCAUGCGAGAAAUCACAUA-GUCAUU--------------UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGA ...(((.((....)).)))(-(((...--------------...)))).((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).)))))))). ( -33.90) >pfo.1282 2549453 104 + 6438405 ACUCCGCGAGAAAUCAAAGAUGUAACC--------------AACGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--AGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGA .......((....))............--------------........(((((((.......((((((.....))))))..--..(((.(..((((....))))..).)))))))))). ( -28.80) >consensus AC_ACGCGAGAAAUCAAAAAUGUAAUC______________UGCGAUUGCUGAGCCAAGUUUAGGGUUUCCUAAAAACCCAA__GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGA .................................................((((((((......((((((.....))))))..........(..((((....))))..)...)))))))). (-25.52 = -25.52 + 0.00)

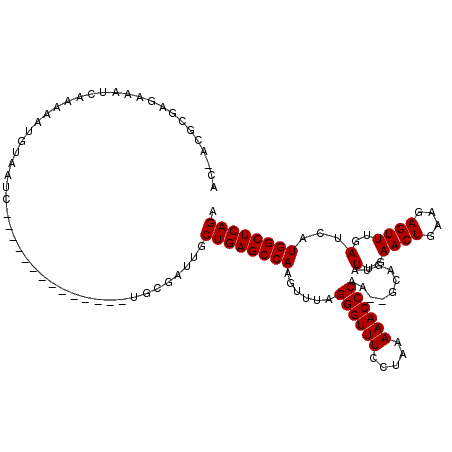
| Location | 722,006 – 722,123 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -24.14 |
| Energy contribution | -23.67 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722006 117 - 6264404 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GU .((((((((.(...........((((.....)))).--..((((((.....))))))....).)))))))).........(((((.....)))))....((.((((......)))).-)) ( -31.40) >pau.2632 1580381 117 + 6537648 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUCUCGAAUUCACGAGU-GU .((((((((.(...........((((.....)))).--..((((((.....))))))....).)))))))).........(((((.....)))))....((.((((......)))).-)) ( -31.40) >pss.496 964606 106 - 6093698 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCUAACUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGUU--------------GGUUACAUCUUUGAUUUCUCGCGGAGU .((((((((.(....((((....)))).............((((((.....))))))....).))))))))...((((.--------------((..(((....)).)..)).))))... ( -23.10) >ppk.94 171296 103 - 6181863 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA--------------AAUGAC-UAUGUGAUUUCUCGCAUGGU .((((((((.(...........((((.....)))).--..(((((((...)))))))....).))))))))........--------------....((-(((((((....))))))))) ( -32.30) >pel.69 115416 103 - 5888780 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA--------------AAUGAC-UAUGUGAUUUCUCGCAUGGC .((((((((.(...........((((.....)))).--..(((((((...)))))))....).))))))))........--------------.....(-(((((((....)))))))). ( -31.10) >pfo.1282 2549453 104 - 6438405 UCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCU--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCGUU--------------GGUUACAUCUUUGAUUUCUCGCGGAGU .((((((((.(....((((....)))).........--..(((((((...)))))))....).))))))))...((((.--------------((..(((....)).)..)).))))... ( -23.50) >consensus UCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC__UUGGGUUUUGAAAAAACCCUAAACUUGGCUCAGCAAUCGCA______________GAUAACAUAUUUGAUUUCUCGCGU_GU .((((((((.(....((((....)))).............(((((((...)))))))....).)))))))).............................((((((......)))))).. (-24.14 = -23.67 + -0.47)

| Location | 722,045 – 722,163 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722045 118 + 6264404 AUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG ...((((..((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))))))..(((((((.((((........).)))..))).)))) ( -39.20) >pau.2632 1580420 118 - 6537648 AUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG ...((((..((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))))))..(((((((.((((........).)))..))).)))) ( -39.20) >pss.496 964633 119 + 6093698 -AACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAGUUAGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG -..((((..(((((((.......((((((.....))))))......(((.(..((((....))))..).))))))))))))))..(((((((.((((........).)))..))).)))) ( -37.40) >ppk.94 171322 117 + 6181863 -UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG -........((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))......(((((((.((((........).)))..))).)))) ( -38.60) >pel.69 115442 117 + 5888780 -UGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG -........((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))......(((((((.((((........).)))..))).)))) ( -38.60) >pfo.1282 2549480 117 + 6438405 -AACGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--AGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG -..((((..(((((((.......((((((.....))))))..--..(((.(..((((....))))..).))))))))))))))..(((((((.((((........).)))..))).)))) ( -37.30) >consensus _UGCGAUUGCUGAGCCAAGUUUAGGGUUUCCUAAAAACCCAA__GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAGGCCUAACACAUGCAAGUCGAGCG .........((((((((......((((((.....))))))..........(..((((....))))..)...))))))))......(((((((.((((........).)))..))).)))) (-35.12 = -35.12 + 0.00)

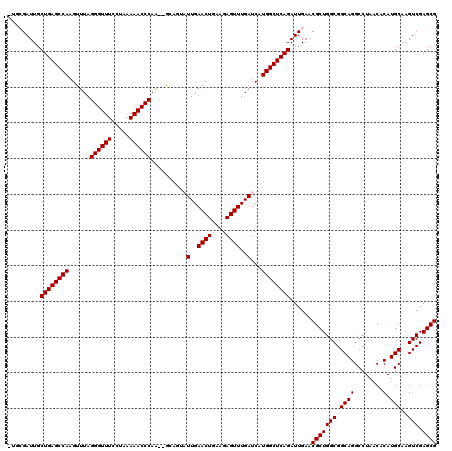
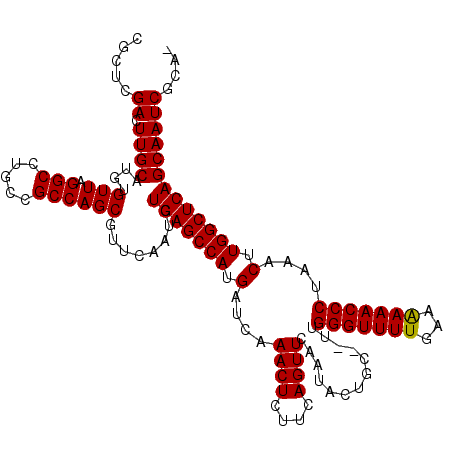
| Location | 722,045 – 722,163 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -28.58 |
| Energy contribution | -28.33 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 722045 118 - 6264404 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAU .(((..((........))..))).........(((......((((((((.(...........((((.....)))).--..((((((.....))))))....).))))))))....))).. ( -32.60) >pau.2632 1580420 118 + 6537648 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAU .(((..((........))..))).........(((......((((((((.(...........((((.....)))).--..((((((.....))))))....).))))))))....))).. ( -32.60) >pss.496 964633 119 - 6093698 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCUAACUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGUU- ....(((.((((....(((.(((.....))))))........(((((((.(....((((....)))).............((((((.....))))))....).))))))))))))))..- ( -29.40) >ppk.94 171322 117 - 6181863 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA- ((((.(((........))).(((.....)))))))......((((((((.(...........((((.....)))).--..(((((((...)))))))....).))))))))........- ( -31.00) >pel.69 115442 117 - 5888780 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCA- ((((.(((........))).(((.....)))))))......((((((((.(...........((((.....)))).--..(((((((...)))))))....).))))))))........- ( -31.00) >pfo.1282 2549480 117 - 6438405 CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCU--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCGUU- ....(((.((((....(((.(((.....))))))........(((((((.(....((((....)))).........--..(((((((...)))))))....).))))))))))))))..- ( -29.80) >consensus CGCUCGACUUGCAUGUGUUAGGCCUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC__UUGGGUUUUGAAAAAACCCUAAACUUGGCUCAGCAAUCGCA_ .....((.((((....(((.(((.....))))))........(((((((.(....((((....)))).............(((((((...)))))))....).))))))))))))).... (-28.58 = -28.33 + -0.25)

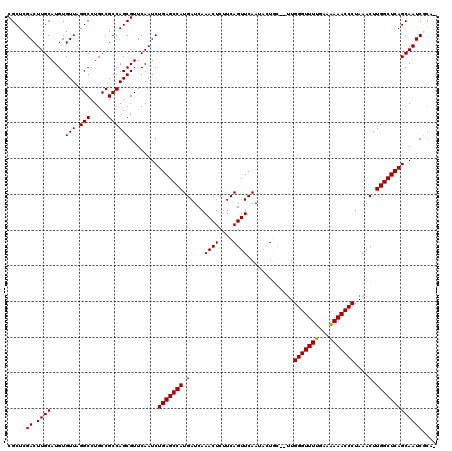
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:07:14 2007