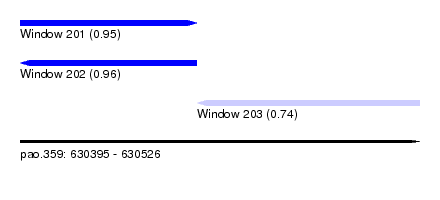
| Sequence ID | pao.359 |
|---|---|
| Location | 630,395 – 630,526 |
| Length | 131 |
| Max. P | 0.962914 |

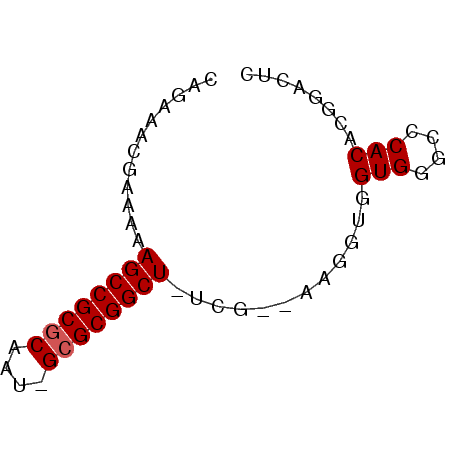
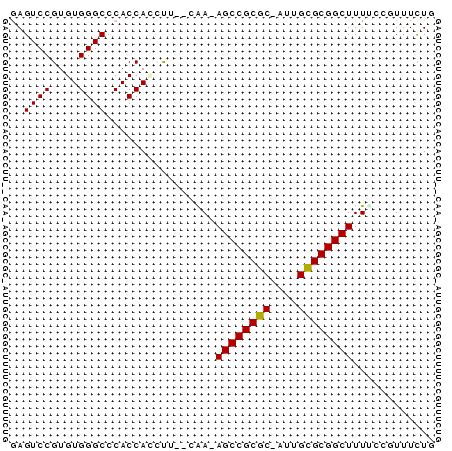
| Location | 630,395 – 630,453 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
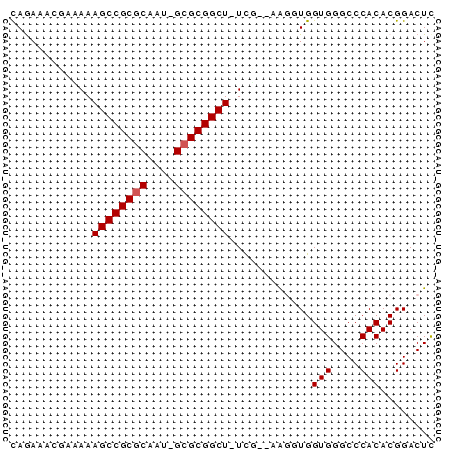
>pao.359 630395 58 + 6264404 UAGAAAGCGCAAAGCCGCGCAGU-GCGCGGCU-UUG--AAGGUGGUGGGCCCACACGGACUC .......(((((((((((((...-))))))))-)))--...((((.....)))).))..... ( -24.80) >pau.363 640368 58 + 6537648 UAGAAAGCGCAAAGCCGCGCAAU-GCGCGGCU-UUG--AAGGUGGUGGGCCCACACGGACUC .......(((((((((((((...-))))))))-)))--...((((.....)))).))..... ( -24.80) >pss.2807 586743 59 - 6093698 CAAAAACAAAAAAGCCGCGCAAU-GCGCGGCUUUCA--AAGGUGGUGGGCCCACACGGACUC ..........((((((((((...-))))))))))..--...((((.....))))........ ( -21.10) >pel.208 431574 58 + 5888780 CAAAAACGCAAAAGCCGCGCAAU-GCGCGGCU-UCG--AAGAUGGUGGGCCCACACGGACUC ......((...(((((((((...-))))))))-)..--......(((....))).))..... ( -19.30) >pf5.3416 619064 59 - 7074893 CAGAAAAGCGAAAGCCGCGCAUU-GCGCGGCUUUCU--UGUUUGGUGGGCCCACACGGACUU .........(((((((((((...-))))))))))).--.((((((((....))).))))).. ( -28.10) >pfo.3036 646722 62 - 6438405 CAGAUACGAAAAAGCCGCACAUAAGCGCGGCUUUUUCAUGUUUGGUGGGCCCACACGGACUU ((((((.(((((((((((.(....).))))))))))).))))))(((....)))........ ( -26.80) >consensus CAGAAACGAAAAAGCCGCGCAAU_GCGCGGCU_UCG__AAGGUGGUGGGCCCACACGGACUC ............((((((((....))))))))............(((....)))........ (-17.82 = -17.98 + 0.17)

| Location | 630,395 – 630,453 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.37 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.359 630395 58 - 6264404 GAGUCCGUGUGGGCCCACCACCUU--CAA-AGCCGCGC-ACUGCGCGGCUUUGCGCUUUCUA (((..(((((((.....))))...--.((-((((((((-...)))))))))))))..))).. ( -26.30) >pau.363 640368 58 - 6537648 GAGUCCGUGUGGGCCCACCACCUU--CAA-AGCCGCGC-AUUGCGCGGCUUUGCGCUUUCUA (((..(((((((.....))))...--.((-((((((((-...)))))))))))))..))).. ( -26.30) >pss.2807 586743 59 + 6093698 GAGUCCGUGUGGGCCCACCACCUU--UGAAAGCCGCGC-AUUGCGCGGCUUUUUUGUUUUUG ......(.((((.....)))))..--.(((((((((((-...)))))))))))......... ( -23.00) >pel.208 431574 58 - 5888780 GAGUCCGUGUGGGCCCACCAUCUU--CGA-AGCCGCGC-AUUGCGCGGCUUUUGCGUUUUUG (((..(((((((.....))))...--.((-((((((((-...)))))))))).)))..))). ( -23.20) >pf5.3416 619064 59 + 7074893 AAGUCCGUGUGGGCCCACCAAACA--AGAAAGCCGCGC-AAUGCGCGGCUUUCGCUUUUCUG ((((..((.(((.....))).)).--.(((((((((((-...)))))))))))))))..... ( -25.90) >pfo.3036 646722 62 + 6438405 AAGUCCGUGUGGGCCCACCAAACAUGAAAAAGCCGCGCUUAUGUGCGGCUUUUUCGUAUCUG ......((.(((.....))).))(((((((((((((((....)))))))))))))))..... ( -28.90) >consensus GAGUCCGUGUGGGCCCACCACCUU__CAA_AGCCGCGC_AUUGCGCGGCUUUUCCGUUUCUG ..((((....))))................((((((((....))))))))............ (-18.50 = -18.37 + -0.14)

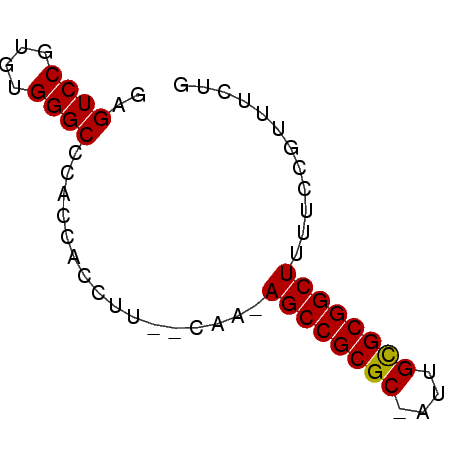
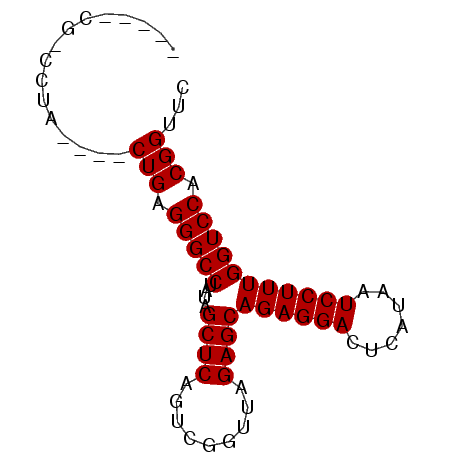
| Location | 630,453 – 630,526 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 87.30 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.359 630453 73 - 6264404 UCCGCCG----ACCUUCUGGGGGCCUAUAGCUCAGUCGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC ...((((----.((....))(((((....((((.........))))((((((.......))))))))))).)))).. ( -24.80) >pau.363 640426 67 - 6537648 ----CCG----AC--UCUGGGGGCCUAUAGCUCAGUCGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC ----(((----((--(....((((.....))))))))))...((.(((((((.......))))))).))........ ( -21.70) >pss.2807 586802 68 + 6093698 -----CGACCUA----CUGAGGGCCCAUAGCUCAGUUGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC -----.((((.(----(((((.........)))))).)))).((.(((((((.......))))))).))........ ( -22.50) >pel.208 431632 66 - 5888780 -------ACCUG----CUGAGGGCCUAUAGCUCAGUCGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC -------....(----(((.(((((....((((.........))))((((((.......))))))))))).)))).. ( -21.90) >pf5.3416 619123 66 + 7074893 -------GCCUA----CUGAGGGCCUAUAGCUCAGUUGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC -------....(----(((.(((((....((((.........))))((((((.......))))))))))).)))).. ( -21.60) >psp.2723 5323393 68 - 5928787 -----CGGCCUA----CUGAGGGCCCAUAGCUCAGUUGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC -----.((((.(----(((((.........)))))).)))).((.(((((((.......))))))).))........ ( -22.00) >consensus _____CG_CCUA____CUGAGGGCCUAUAGCUCAGUCGGUUAGAGCAGAGGACUCAUAAUCCUUUGGUCCACGGUUC ................(((.(((((....((((.........))))((((((.......))))))))))).)))... (-20.10 = -20.10 + -0.00)

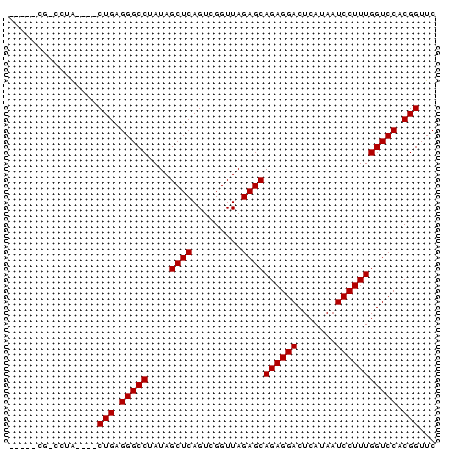
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:06:53 2007