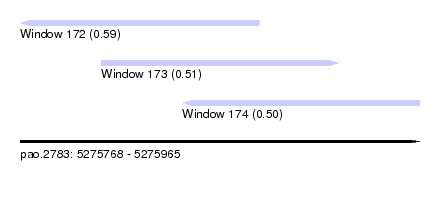
| Sequence ID | pao.2783 |
|---|---|
| Location | 5,275,768 – 5,275,965 |
| Length | 197 |
| Max. P | 0.592409 |

| Location | 5,275,768 – 5,275,886 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -24.35 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
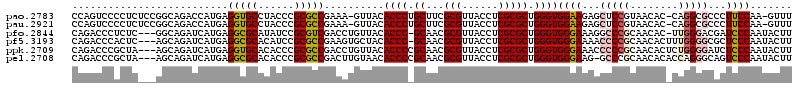
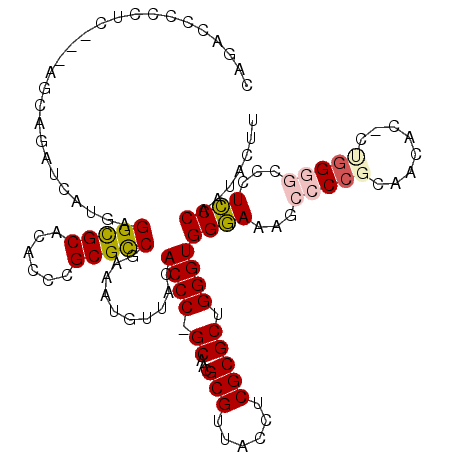
>pao.2783 5275768 118 - 6264404 UGGAAGAGCUCCGUAACAC-CAGGCGCCCUUCCAA-GUUUGCUCAAUCGGCAGUUGACGUAGCCGGUGACUACCGUCACCCAUGCUCUGCCGGGCAGCCAACCGAAUUUCGGUCGCCCGC ((((((.((.((.......-..)).)).)))))).-....((......(((((.....(((...((((((....))))))..))).)))))((((.....((((.....)))).)))))) ( -44.50) >psp.505 4886577 117 + 5928787 UGGGAAUGUUCCGCAACAC-UCAGGGACCUCCCAAUACUUGCUCAGUCAGU-GCUGACGUAGUCGGCGACCACCGUCGCUCAUGCUCUGCUG-GCAGUAAACCUAUUAAGACCCGUCCCG .((((..(((.........-...(((....)))..((((.((.(((.(((.-(((((((....))(((((....)))))))).)).))))))-))))))..........)))...)))). ( -37.00) >pfo.2844 5404857 118 - 6438405 UGGGAAAGGCCCGCAACAC-UUGGGACGAUCCCAAUACUUGCUCAGUCAGU-GCUGACGUAGUCGGCGACCACCGUCGCUCAUGCUCUGCCGAGAAGUAAACCUAUUAAGACCCGUUCGC .(((.....)))(((((..-(((((.....)))))(((((.(((.(.(((.-(((((((....))(((((....)))))))).)).)))).))))))))...............))).)) ( -35.40) >pf5.3193 6020698 119 - 7074893 UGGGAAAACCCCGCAACACUUUGGGGCGCUCCCAAUACUUGCUCAGUCAGU-GCUGACGUAGUCGGCGACCACCGUCGCUCAUGCUCUGCUGAGAAGUAAACCUAUUAAGACCCGCUCCC (((((...(((((........)))))...))))).(((((.(((((.(((.-(((((((....))(((((....)))))))).)).)))))))))))))..................... ( -41.80) >ppk.2709 5319407 104 - 6181863 UGGGAAACCCCCGCAACACUCUGGGGAUCUCCCAAUACUUGCUCAGUCAGU-GCUGACGU---------------UCGCUCAUGCUCUGCUUGGCAGUAAACCUAUUAAGACCCGUCCAG (((((...(((((........)))))...))))).((((.(((.((.(((.-(((((.(.---------------...)))).)).))))).)))))))..................... ( -30.20) >pss.492 5135564 117 + 6093698 UGGGAAUGUUCCGCAACAC-UCAGGGACCUCCCAAUACUUGCUCAGUCAGU-GCUGACGUAGUCGGCGACCACCGUCGCUCAUGCUCUGCUG-GCAGUAAACCUAUUAAGACCCGUCCCG .((((..(((.........-...(((....)))..((((.((.(((.(((.-(((((((....))(((((....)))))))).)).))))))-))))))..........)))...)))). ( -37.00) >consensus UGGGAAAGCCCCGCAACAC_UUGGGGACCUCCCAAUACUUGCUCAGUCAGU_GCUGACGUAGUCGGCGACCACCGUCGCUCAUGCUCUGCUG_GCAGUAAACCUAUUAAGACCCGUCCCC (((((...(((((........)))))...))))).((((.((((.(((((...)))))(((...((((((....))))))..)))......))))))))..................... (-24.35 = -25.48 + 1.13)

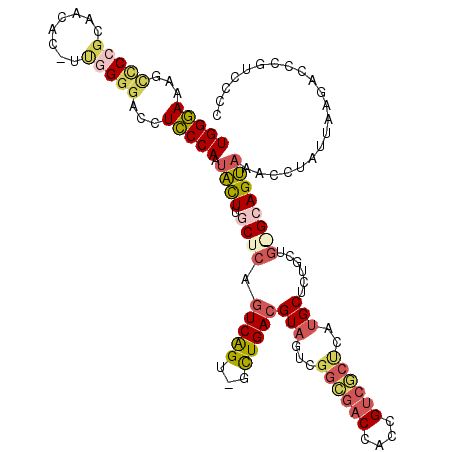
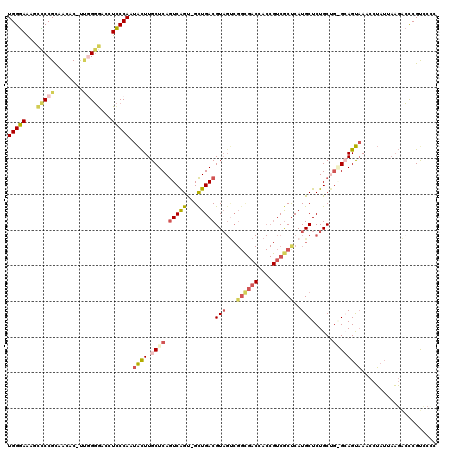
| Location | 5,275,808 – 5,275,925 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -45.45 |
| Consensus MFE | -30.58 |
| Energy contribution | -32.28 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2783 5275808 117 + 6264404 GGUGACGGUAGUCACCGGCUACGUCAACUGCCGAUUGAGCAAAC-UUGGAAGGGCGCCUG-GUGUUACGGAGCUCUUCCACCCAGCGCGAGGUAACGCGAAGCAGGGUGUAAC-UUUCGG ((((((....))))))(((..........)))((..(.......-..((((((((..(((-......))).))))))))((((..((((......)))).....))))....)-..)).. ( -43.90) >pst.577 5331612 117 - 6397126 AGCGACGGUGGUCGCCGACUACGUCAGC-ACUGACUGAGCAAGUAUUGGGAAGUCCCUGA-GUGUUGCGGAACAUUCCCACCCAGCGCGAGGUAACGCGUUGC-GGGUGUUACAGGUCGG .((((((.(((((...))))))))).))-.((((((.((((.....((((((.((((...-.....).)))...))))))(((((((((......))))))).-)).))))...)))))) ( -46.80) >pfo.2844 5404897 117 + 6438405 AGCGACGGUGGUCGCCGACUACGUCAGC-ACUGACUGAGCAAGUAUUGGGAUCGUCCCAA-GUGUUGCGGGCCUUUCCCACCCAGCGCGAGGUAACGCGUUGC-GGGUGUAACAGGUCGA .(((((....)))))......((.((((-(((.(((.....)))..((((.....)))))-))))))))(((((.(..(((((((((((......))))))..-)))))..).))))).. ( -48.70) >pf5.3193 6020738 118 + 7074893 AGCGACGGUGGUCGCCGACUACGUCAGC-ACUGACUGAGCAAGUAUUGGGAGCGCCCCAAAGUGUUGCGGGGUUUUCCCACCCAGCGCGAGGUAACGCGUUGC-GGGUGUAGCACUUCGG ......((((.((.((((.(((.((((.-.....))))....))))))))).))))((.(((((((((((((....))).(((((((((......))))))).-)).)))))))))).)) ( -49.00) >ppk.2709 5319447 104 + 6181863 AGCGA---------------ACGUCAGC-ACUGACUGAGCAAGUAUUGGGAGAUCCCCAGAGUGUUGCGGGGGUUUCCCACCCAGCGCGAGGUAACGCGUUGCGGGGUGUAACAGGUCGG .((..---------------......))-.((((((..(((.....((((((((((((..........))))))))))))(((((((((......))))))).))..)))....)))))) ( -44.50) >pel.2708 5010528 103 + 5888780 AGCGA---------------ACGUCAGC-ACUGACUGAGCAAGUAUUGGGACUGCCCUGGUGUGUUGCGGGC-CUUCCCACCCAGCGCGAGGUAACGCGUUGCGGGGUGUUACAAGUCGG .((..---------------......))-.((((((.((((.....(((((..((((...........))))-..)))))(((((((((......))))))).))..))))...)))))) ( -39.80) >consensus AGCGACGGUGGUCGCCGACUACGUCAGC_ACUGACUGAGCAAGUAUUGGGAGCGCCCCAA_GUGUUGCGGGGCUUUCCCACCCAGCGCGAGGUAACGCGUUGC_GGGUGUAACAGGUCGG .((((((.(((((...))))))))).))..((((((.........(((((.....)))))..(((((((((.....)))((((((((((......))))))...)))))))))))))))) (-30.58 = -32.28 + 1.70)

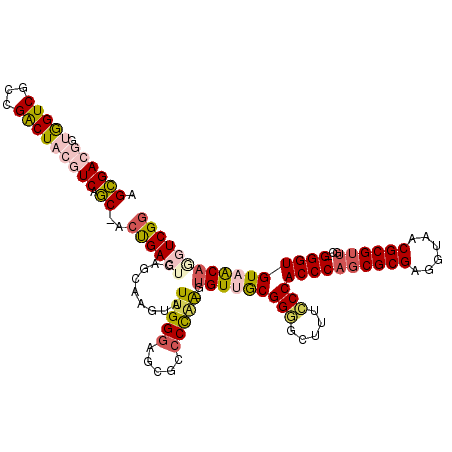
| Location | 5,275,848 – 5,275,965 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>pao.2783 5275848 117 - 6264404 CCAGUCCCCUCUCCGGCAGACCAUGAGGUGCCUACCCGCGCCGAAA-GUUACACCCUGCUUCGCGUUACCUCGCGCUGGGUGGAAGAGCUCCGUAACAC-CAGGCGCCCUUCCAA-GUUU ..((.(.((((...((....))..)))).).))....(((((....-(((((((((.((...(((......))))).))))(((.....))))))))..-..)))))........-.... ( -36.40) >pau.2921 5547780 117 - 6537648 CCAGUCCCCUCUCCGGCAGACCAUGAGGUGCCUACCCGCGCCGAAA-GUUACACCCUGCUUCGCGUUACCUCGCGCUGGGUGGAAGAGCUCCGUAACAC-CAGGCGCCCUUCCAA-GUUU ..((.(.((((...((....))..)))).).))....(((((....-(((((((((.((...(((......))))).))))(((.....))))))))..-..)))))........-.... ( -36.40) >pfo.2844 5404936 115 - 6438405 CAGACCCUCUC---GGCAGAUCAUGAGGCGCAUAUCCGCGUCGACCUGUUACACCC-GCAACGCGUUACCUCGCGCUGGGUGGGAAAGGCCCGCAACAC-UUGGGACGAUCCCAAUACUU ..(..(((.((---(((((.((....(((((......))))))).))))).(((((-(...((((......)))).)))))).)).)))..).......-(((((.....)))))..... ( -36.80) >pf5.3193 6020777 116 - 7074893 CAGACCCACUC---AGCAGAUCAUGAGGCGCACAUCCGCGCCGAAGUGCUACACCC-GCAACGCGUUACCUCGCGCUGGGUGGGAAAACCCCGCAACACUUUGGGGCGCUCCCAAUACUU ....(((((((---(((....(((..(((((......)))))...)))........-.....(((......)))))))))))))....(((((........))))).............. ( -43.60) >ppk.2709 5319471 117 - 6181863 CAGACCCGCUA---AGCAGAUCAUGAGGUGCACACCCGCGCCGACCUGUUACACCCCGCAACGCGUUACCUCGCGCUGGGUGGGAAACCCCCGCAACACUCUGGGGAUCUCCCAAUACUU ((((.......---(((((.((....(((((......))))))).))))).......((..((((......))))..(((.((....))))))).....))))(((....)))....... ( -39.30) >pel.2708 5010552 116 - 5888780 CAGACCCGCUA---AGCAGAUCAUGAGGCGCACACCCGCGCCGACUUGUAACACCCCGCAACGCGUUACCUCGCGCUGGGUGGGAAG-GCCCGCAACACACCAGGGCAGUCCCAAUACUU ...((((((..---.......((...(((((......)))))....)).........))..((((......))))..))))((((..-((((...........))))..))))....... ( -40.27) >consensus CAGACCCCCUC___AGCAGAUCAUGAGGCGCACACCCGCGCCGAAAUGUUACACCC_GCAACGCGUUACCUCGCGCUGGGUGGGAAAGCCCCGCAACAC_CUGGGGCCCUCCCAAUACUU ..........................(((((......)))))..........((((.((...(((......))))).))))((((...(((((........)))))...))))....... (-27.86 = -28.12 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:06:23 2007