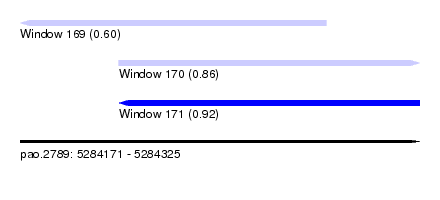
| Sequence ID | pao.2789 |
|---|---|
| Location | 5,284,171 – 5,284,325 |
| Length | 154 |
| Max. P | 0.916167 |

| Location | 5,284,171 – 5,284,289 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.96 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2789 5284171 118 - 6264404 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCUUGG-UCUCUCAGCUUA-CCUGCUGGCCUCGCGACCAGUUGGUGUAAUAAUAACUAUUCUCAUUAUCAGGUCAA ...........(((((((((.((((....((((.(((.((((....))))-.)))))))..((-((.(((((........))))).)))).............)))).)))))))))... ( -39.60) >pau.2927 5556103 118 - 6537648 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCUUGG-UCUCUCAGCUUA-CCUGCUGGCCUCGCGACCAGUUGGUGUAAUAAUAACUAUUCUCAUUAUCAGGUCAA ...........(((((((((.((((....((((.(((.((((....))))-.)))))))..((-((.(((((........))))).)))).............)))).)))))))))... ( -39.60) >pel.2712 5016965 118 - 5888780 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAG-GGAGCUUUCAGGUUACCCAACCAGUCUCGCGACCAGCUGUGAUAAUCAUAACGAUUCUCAUUACGUAGUCAA .....((((((.(.(((((((((..((((((((-((((((.......-))).)))))))))))........(((.(((.....)).).)))...........))))))))).))))))). ( -33.40) >ppk.2712 5325378 117 - 6181863 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAG-GG-GCUCUUAGCUUAUCCAACCAGUCUCGCGACCAGUUGCGAUAAUAAUAACAAUUCUCAUUUAGCCGUCAA .....((((..((..(((((((((((((.((((-((((((.......-))-)))))))).))))).........((((((....))))))............))))))))..)).)))). ( -39.80) >pf5.3196 6028152 117 - 7074893 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCACUGG-UCUUCCAGGUUA-UCGACUGAUCUCGCGACCAGUUGUGAUAAUCAUAACGAUUCUCAUUUCGAAGUCAA ...........((..(((.((..(((((((((.-.((.(((......)))-.))..)))))))-))..)).)))..))(((..(..((((.((((.....)))).))))..)...))).. ( -38.40) >pfo.2848 5412549 117 - 6438405 AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAAGGG-CUUUUCAGAAUA-UCGACUGAUCUCGCGAUCAGUUGUGAUAAUCAUAGCGAUUCUCAUUACCAAGUCAA .....(((((....(((((((((((((.(((((-((((((.......)))-).))))))).))-))((((((((....))))))))................)))))))))...))))). ( -39.40) >consensus AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA_GAGUCCGAACCAGUGG_UCUCUCAGCUUA_CCAACUGGUCUCGCGACCAGUUGUGAUAAUAAUAACGAUUCUCAUUACCAAGUCAA .....(((((....(((((((((......((((.(((...............)))))))......(((((((((....)))))))))...............)))))))))...))))). (-19.82 = -20.96 + 1.14)
| Location | 5,284,209 – 5,284,325 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -44.15 |
| Consensus MFE | -33.04 |
| Energy contribution | -32.02 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
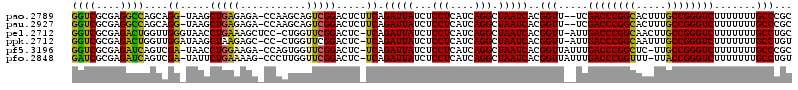
>pao.2789 5284209 116 + 6264404 GGUCGCGAGGCCAGCAGG-UAAGCUGAGAGA-CCAAGCAGUCGGACUCUUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU--UCGACCCGGCACUUUGCCGGGUCUUUUUUUGCCCGC ((((....)))).((.((-((((....((((-(((((.((.....)))))..(((((...(((....))).)))))..))))--))((((((((.....))))))))....)))))).)) ( -46.30) >pau.2927 5556141 116 + 6537648 GGUCGCGAGGCCAGCAGG-UAAGCUGAGAGA-CCAAGCAGUCGGACUCUUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU--UCGACCCGGCACUUUGCCGGGUCUUUUUUUGCCCGC ((((....)))).((.((-((((....((((-(((((.((.....)))))..(((((...(((....))).)))))..))))--))((((((((.....))))))))....)))))).)) ( -46.30) >pel.2712 5017003 117 + 5888780 GGUCGCGAGACUGGUUGGGUAACCUGAAAGCUCC-CUGGUUCGGACUC-UCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU-AUUGACCCGGCAACUUGCCGGGUCUUUUUUUGCCUGC ((((....))))((..((((((.((((.((.(((-.......))))).-)))).))))))))....(((((((((....)))-)..((((((((.....)))))))).......))))). ( -45.00) >ppk.2712 5325416 116 + 6181863 GGUCGCGAGACUGGUUGGAUAAGCUAAGAGC-CC-CUGGUUCGGACUC-UCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU-AUUGACCCGGCAAUUUGCCGGGUCUUUUUUUGCCUGU ((((....))))((..((((((.((.(((((-(.-.......)).)))-).)).))))))))....(((((((((....)))-)..((((((((.....)))))))).......))))). ( -44.90) >pf5.3196 6028190 116 + 7074893 GGUCGCGAGAUCAGUCGA-UAACCUGGAAGA-CCAGUGGUUCGGACUC-UCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUUUGACCCGGCUC-UUGCCGGGUCUUUUUUUGCCCGC ....(((.(((.((..((-(((.(((..((.-((........)).)).-.))).)))))..)).))).(((((((....))))...((((((((..-..)))))))).......)))))) ( -41.00) >pfo.2848 5412587 116 + 6438405 GAUCGCGAGAUCAGUCGA-UAUUCUGAAAAG-CCCUUGGUUCGGACUC-UCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUUUGACCCGGUUU-UUACCGGGUCUUUUUUUGCCUGU ((((....))))((..((-((.(((((..((-.((.......)).)).-))))).))))..))...(((((((((....))))...((((((((..-..)))))))).......))))). ( -41.40) >consensus GGUCGCGAGACCAGUAGG_UAAGCUGAAAGA_CCAAUGGUUCGGACUC_UCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU__UUGACCCGGCACUUUGCCGGGUCUUUUUUUGCCCGC ((((....))))....((.....(((((...........))))).....)).(((((...(((....))).)))))..(((.....((((((((.....)))))))).......)))... (-33.04 = -32.02 + -1.02)

| Location | 5,284,209 – 5,284,325 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -47.53 |
| Consensus MFE | -29.28 |
| Energy contribution | -28.34 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
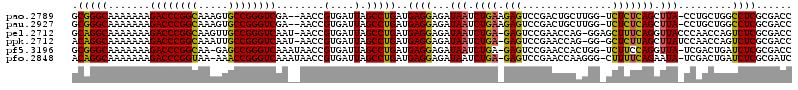
>pao.2789 5284209 116 - 6264404 GCGGGCAAAAAAAGACCCGGCAAAGUGCCGGGUCGA--AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCUUGG-UCUCUCAGCUUA-CCUGCUGGCCUCGCGACC ((((((.......((((((((.....))))))))..--......(((((.(((....)))...)))))...((((.((((....))))-.))))((((...-...))))))).))).... ( -48.00) >pau.2927 5556141 116 - 6537648 GCGGGCAAAAAAAGACCCGGCAAAGUGCCGGGUCGA--AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCUUGG-UCUCUCAGCUUA-CCUGCUGGCCUCGCGACC ((((((.......((((((((.....))))))))..--......(((((.(((....)))...)))))...((((.((((....))))-.))))((((...-...))))))).))).... ( -48.00) >pel.2712 5017003 117 - 5888780 GCAGGCAAAAAAAGACCCGGCAAGUUGCCGGGUCAAU-AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAG-GGAGCUUUCAGGUUACCCAACCAGUCUCGCGACC ((((((.......((((((((.....))))))))..(-((((..(((((.(((....)))...)))))(((-((((((.......-))).)))))))))))........)))).)).... ( -45.40) >ppk.2712 5325416 116 - 6181863 ACAGGCAAAAAAAGACCCGGCAAAUUGCCGGGUCAAU-AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAG-GG-GCUCUUAGCUUAUCCAACCAGUCUCGCGACC .(((((.......((((((((.....))))))))..(-((......))))))))....((.(((((.((((-((((((.......-))-)))))))).)))))...)).(((....))). ( -48.20) >pf5.3196 6028190 116 - 7074893 GCGGGCAAAAAAAGACCCGGCAA-GAGCCGGGUCAAAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCACUGG-UCUUCCAGGUUA-UCGACUGAUCUCGCGACC ((((((.......((((((((..-..))))))))...(((......)))))).(((.((..(((((((((.-.((.(((......)))-.))..)))))))-))..)).))).))).... ( -50.40) >pfo.2848 5412587 116 - 6438405 ACAGGCAAAAAAAGACCCGGUAA-AAACCGGGUCAAAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA-GAGUCCGAACCAAGGG-CUUUUCAGAAUA-UCGACUGAUCUCGCGAUC .(((((.......((((((((..-..))))))))...(((......))))))))...((..((((.(((((-((((((.......)))-).))))))).))-))..))((((....)))) ( -45.20) >consensus GCAGGCAAAAAAAGACCCGGCAAAGUGCCGGGUCAA__AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGA_GAGUCCGAACCAGUGG_UCUCUCAGCUUA_CCAACUGGUCUCGCGACC .(((((.......((((((((.....))))))))........(....).)))))..((((...(((.((((.(((...............))))))).))).........))))...... (-29.28 = -28.34 + -0.94)
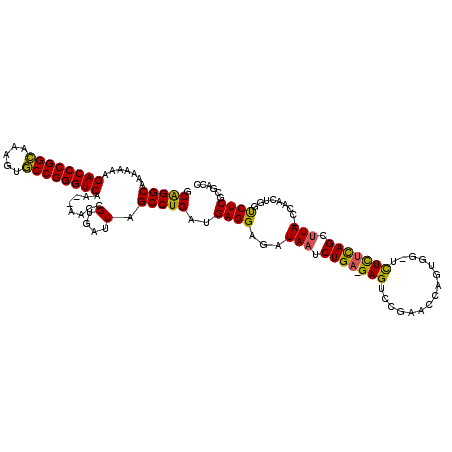
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:06:20 2007