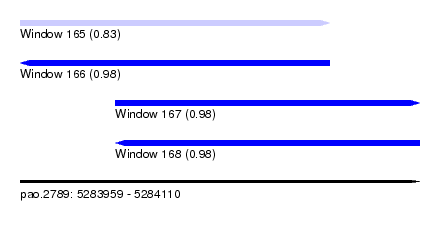
| Sequence ID | pao.2789 |
|---|---|
| Location | 5,283,959 – 5,284,110 |
| Length | 151 |
| Max. P | 0.981443 |

| Location | 5,283,959 – 5,284,076 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2789 5283959 117 + 6264404 UUGACAUGGAAAUGAGAAUCAUUAUUAUGUCACUCAACUGGUCGCGAGA-UCAGCCGGUAAGCUGAGA--GACCCACGCAGUCGGACUCUUCAGAUUAUCUCCUCAUCAGGCUAAUCACG .((((((((.((((.....)))).)))))))).....((((((....))-))))..(((((.((((((--(.((.((...)).)).))).)))).))))).(((....)))......... ( -33.20) >pf5.2389 4555422 114 + 7074893 UUGACUUGCAAAUGAUAAUGAUUAUUAUUGCAAGCAACUGGUCGCGAGA-UCAGUCGAUAGCCCAAAG-GACCUUA---GGUCGGA-CCUUUGGAUUAUCUCCUCAUCAGGCUAAUCACG .((.(((((((.(((((.....))))))))))))))(((((((....))-))))).((((..((((((-(.((...---....)).-)))))))..)))).(((....)))......... ( -38.00) >pss.1813 3620502 112 + 6093698 UUGACUUGCAAAUGAUAAUGAUUAUUAUUGC-AUCAGCGGGUCGCGAGACUCGCUGGAUAACCCGGAA--GUCUUA---GGUCGG--CUUCCAGAUUAUCUCCUCAUCAGGCUAAUCACG ......(((((.(((((.....)))))))))-)((((((((((....)))))))))).......((((--(((...---....))--))))).(((((...(((....))).)))))... ( -39.20) >pel.1954 3656602 116 + 5888780 UUGACUGGCAAAUGAUAACGAUUAUUAUUGCACUCAGCAGGUCGCGAGACCCGCUGGAUAACCUGCAACGUCCUCA---GGUCGGA-CGCUCAGAUUAUCUCCUCAUCAGGCUAAUCACG .(((.((((...((((..................((((.((((....)))).))))(((((.(((...(((((...---....)))-))..))).))))).....)))).)))).))).. ( -38.90) >ppk.2347 4595202 111 + 6181863 UUGACUGCCAAAUGAUAACGAUUAUUAUUGCACUCAGCUGAUCGCGAGAUCCGCUGGAUAACCUGAAA---GCUUA---GGUCG---CUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACG .(((..(((..((((...................((((.((((....)))).))))(((((.((((.(---((...---....)---)).)))).)))))...))))..)))...))).. ( -34.00) >pfo.2147 4145110 113 + 6438405 UUGACUUGCAAAUGAUAAUGAUUAUUAUUGCAAGCAACUGGUCGCGAGA-UCAGUCGAUGGACCAGAG--ACCUUA---GGUCGGU-CUUCUGGACUAUCUCCUCAUCAGGCUAAUCACG .((.(((((((.(((((.....))))))))))))))(((((((....))-))))).(((((.((((((--(((...---....)))-).))))).))))).(((....)))......... ( -40.40) >consensus UUGACUUGCAAAUGAUAAUGAUUAUUAUUGCAAUCAACUGGUCGCGAGA_UCACUCGAUAACCCGAAA__ACCUUA___GGUCGGA_CUUUCAGAUUAUCUCCUCAUCAGGCUAAUCACG .(((...((...((((.......................((((....)).))....(((((.((((((...((..........))...)))))).))))).....)))).))...))).. (-14.22 = -14.00 + -0.22)

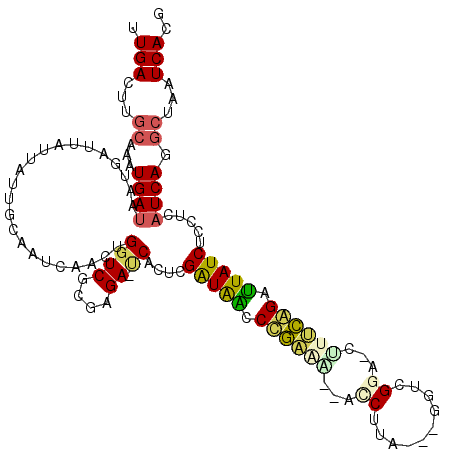
| Location | 5,283,959 – 5,284,076 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2789 5283959 117 - 6264404 CGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCGUGGGUC--UCUCAGCUUACCGGCUGA-UCUCGCGACCAGUUGAGUGACAUAAUAAUGAUUCUCAUUUCCAUGUCAA ...(((((.(((....)))...))))).........((((((((((((..--..((((((....))))))-.))))))..))))))..((((((...((((.....))))...)))))). ( -35.40) >pf5.2389 4555422 114 - 7074893 CGUGAUUAGCCUGAUGAGGAGAUAAUCCAAAGG-UCCGACC---UAAGGUC-CUUUGGGCUAUCGACUGA-UCUCGCGACCAGUUGCUUGCAAUAAUAAUCAUUAUCAUUUGCAAGUCAA (((((.......(((.((..((((..(((((((-.((....---...)).)-))))))..))))..)).)-))))))).......((((((((..(((.....)))...))))))))... ( -38.81) >pss.1813 3620502 112 - 6093698 CGUGAUUAGCCUGAUGAGGAGAUAAUCUGGAAG--CCGACC---UAAGAC--UUCCGGGUUAUCCAGCGAGUCUCGCGACCCGCUGAU-GCAAUAAUAAUCAUUAUCAUUUGCAAGUCAA ..(((((.((.((((((.(((((((((((((((--.(....---...).)--))))))))))))(((((.(((....))).)))))..-..........)).))))))...)).))))). ( -40.30) >pel.1954 3656602 116 - 5888780 CGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGCG-UCCGACC---UGAGGACGUUGCAGGUUAUCCAGCGGGUCUCGCGACCUGCUGAGUGCAAUAAUAAUCGUUAUCAUUUGCCAGUCAA ..(((((.((.((((((.(((((((((((((((-(((....---...))))))).)))))))))(((((((((....))))))))).............)).))))))...)).))))). ( -51.50) >ppk.2347 4595202 111 - 6181863 CGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAG---CGACC---UAAGC---UUUCAGGUUAUCCAGCGGAUCUCGCGAUCAGCUGAGUGCAAUAAUAAUCGUUAUCAUUUGGCAGUCAA ..(((((.(((((((((.(((((((((((((((---(....---...))---))))))))))))((((.((((....)))).)))).............)).))))))...)))))))). ( -45.80) >pfo.2147 4145110 113 - 6438405 CGUGAUUAGCCUGAUGAGGAGAUAGUCCAGAAG-ACCGACC---UAAGGU--CUCUGGUCCAUCGACUGA-UCUCGCGACCAGUUGCUUGCAAUAAUAAUCAUUAUCAUUUGCAAGUCAA (((((.......(((.((..(((.(.(((((.(-(((....---...)))--)))))).).)))..)).)-))))))).......((((((((..(((.....)))...))))))))... ( -35.81) >consensus CGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAAG_UCCGACC___UAAGGC__UUUCAGGUUAUCCACCGA_UCUCGCGACCAGCUGAGUGCAAUAAUAAUCAUUAUCAUUUGCAAGUCAA ..(((((.((.((((((.((((((((((((((....(..........)....))))))))))))...........((.....))...............)).))))))...)).))))). (-18.10 = -18.35 + 0.25)

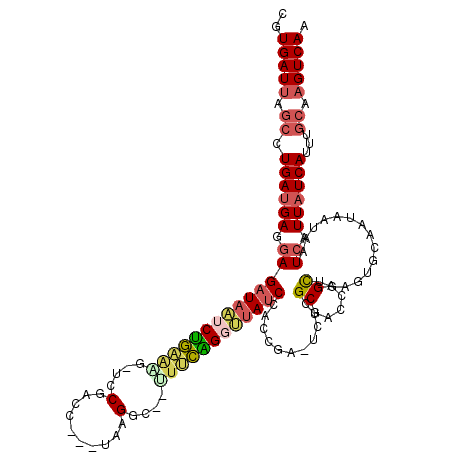
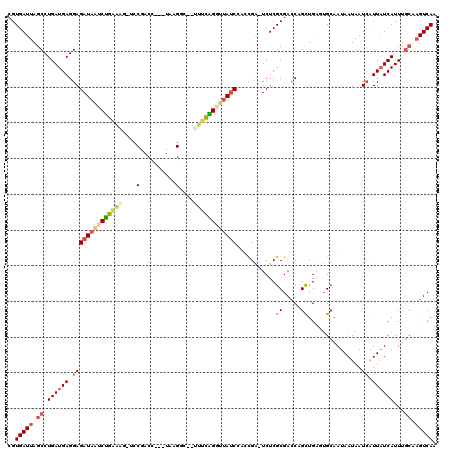
| Location | 5,283,995 – 5,284,110 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -28.99 |
| Energy contribution | -27.72 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.53 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2789 5283995 115 + 6264404 ACUGGUCGCGAGA-UCAGCCGGUAAGCUGAGA--GACCCACGCAGUCGGACUCUUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGC-ACUUUGCCGGGUC-UUUUUUU .((((((....))-))))(((((((.((((((--(.((.((...)).)).))).)))).))))).(((....)))........)).....((((((((-.....))))))))-....... ( -41.90) >pf5.2389 4555458 112 + 7074893 ACUGGUCGCGAGA-UCAGUCGAUAGCCCAAAG-GACCUUA---GGUCGGA-CCUUUGGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGC-UUUUUGCCGGGUC-UUUUUUU (((((((....))-))))).((((..((((((-(.((...---....)).-)))))))..))))........(((((......)))))..((((((((-.....))))))))-....... ( -45.30) >pst.1877 3549491 112 + 6397126 GCGGUUCGCGAGAACCGCUGGAUAACCUGGAA--GUCUUA---GGUCGG--CUUCCGGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGA-UUUUUUCCGGGUCUUUUUUUU (((((((....))))))).((((((.((((((--(((...---....))--))))))).)))))).......(((((......)))))..((((((((-.....))))))))........ ( -49.60) >pel.1954 3656638 114 + 5888780 GCAGGUCGCGAGACCCGCUGGAUAACCUGCAACGUCCUCA---GGUCGGA-CGCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGC-UUUUUGCCGGGUC-UUUUUUU ((.((((....)))).)).((((((.(((...(((((...---....)))-))..))).)))))).......(((((......)))))..((((((((-.....))))))))-....... ( -43.80) >ppk.2347 4595238 110 + 6181863 GCUGAUCGCGAGAUCCGCUGGAUAACCUGAAA---GCUUA---GGUCG---CUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGCUUUUUUGCCGGGUC-UUUUUUU ((.((((....)))).)).((((((.((((.(---((...---....)---)).)))).)))))).......(((((......)))))..((((((((......))))))))-....... ( -40.20) >pfo.2147 4145146 111 + 6438405 ACUGGUCGCGAGA-UCAGUCGAUGGACCAGAG--ACCUUA---GGUCGGU-CUUCUGGACUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGC-UUUUUGCCGGGUC-UUUUUUU (((((((....))-))))).(((((.((((((--(((...---....)))-).))))).)))))........(((((......)))))..((((((((-.....))))))))-....... ( -47.70) >consensus ACUGGUCGCGAGA_CCACUCGAUAACCUGAAA__ACCUUA___GGUCGGA_CUUUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUUUUGACCCGGC_UUUUUGCCGGGUC_UUUUUUU ...((((....)))).....(((((.((((((...((..........))...)))))).)))))........(((((......)))))..((((((((......))))))))........ (-28.99 = -27.72 + -1.27)

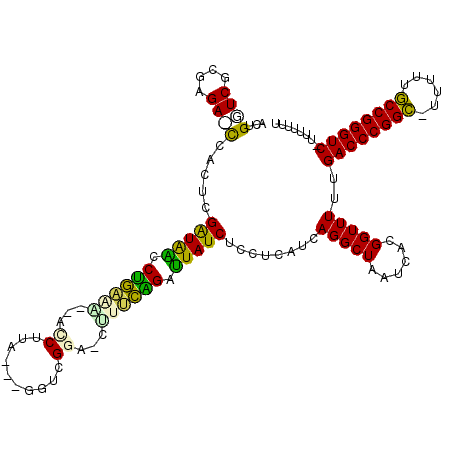
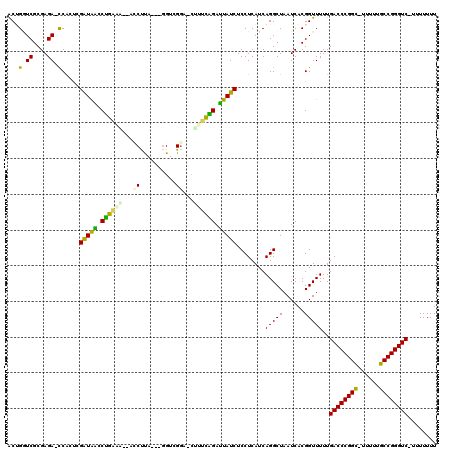
| Location | 5,283,995 – 5,284,110 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -47.30 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.08 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -5.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2789 5283995 115 - 6264404 AAAAAAA-GACCCGGCAAAGU-GCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGACUGCGUGGGUC--UCUCAGCUUACCGGCUGA-UCUCGCGACCAGU .......-((((((((.....-)))))))).....(((.((((((((.(.((((........((((.(((.((.((...)).)).)--))))))))))).))))))-)).)).)...... ( -45.40) >pf5.2389 4555458 112 - 7074893 AAAAAAA-GACCCGGCAAAAA-GCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCCAAAGG-UCCGACC---UAAGGUC-CUUUGGGCUAUCGACUGA-UCUCGCGACCAGU .......-((((((((.....-))))))))......(((((.......(((.((..((((..(((((((-.((....---...)).)-))))))..))))..)).)-)))))))...... ( -49.31) >pst.1877 3549491 112 - 6397126 AAAAAAAAGACCCGGAAAAAA-UCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCCGGAAG--CCGACC---UAAGAC--UUCCAGGUUAUCCAGCGGUUCUCGCGAACCGC ........((((((((.....-))))))))...(((((.(((((.(((....)))...))))))(((((--.(....---...).)--)))).)))).....(((((((....))))))) ( -43.80) >pel.1954 3656638 114 - 5888780 AAAAAAA-GACCCGGCAAAAA-GCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGCG-UCCGACC---UGAGGACGUUGCAGGUUAUCCAGCGGGUCUCGCGACCUGC .......-((((((((.....-))))))))...............(((....))).(((((((((((((-(((....---...))))))).)))))))))..(((((((....))))))) ( -52.80) >ppk.2347 4595238 110 - 6181863 AAAAAAA-GACCCGGCAAAAAAGCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAG---CGACC---UAAGC---UUUCAGGUUAUCCAGCGGAUCUCGCGAUCAGC .......-((((((((......))))))))........(((((..(((....))).(((((((((((((---(....---...))---))))))))))))..(((.....)))))))).. ( -46.20) >pfo.2147 4145146 111 - 6438405 AAAAAAA-GACCCGGCAAAAA-GCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAGUCCAGAAG-ACCGACC---UAAGGU--CUCUGGUCCAUCGACUGA-UCUCGCGACCAGU .......-((((((((.....-))))))))......(((((.......(((.((..(((.(.(((((.(-(((....---...)))--)))))).).)))..)).)-)))))))...... ( -46.31) >consensus AAAAAAA_GACCCGGCAAAAA_GCCGGGUCAAAAACCGUGAUUAGCCUGAUGAGGAGAUAAUCCGAAAG_UCCGACC___UAAGGC__UUUCAGGUUAUCCACCGA_UCUCGCGACCAGC ........((((((((......))))))))......(((((....(((....))).((((((((((((....(..........)....)))))))))))).........)))))...... (-33.38 = -33.08 + -0.30)
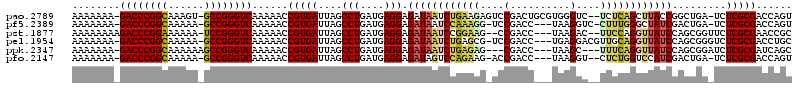
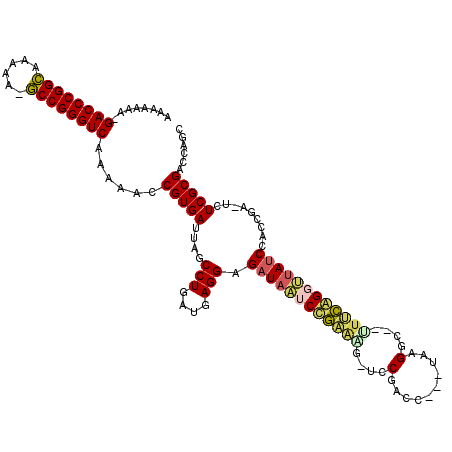
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:06:17 2007