| Sequence ID | pao.3275 |
|---|---|
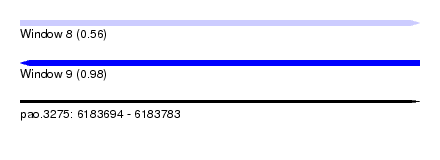
| Location | 6,183,694 – 6,183,783 |
| Length | 89 |
| Max. P | 0.978755 |

| Location | 6,183,694 – 6,183,783 |
|---|---|
| Length | 89 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 43.32 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -4.19 |
| Energy contribution | -6.25 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560546 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.3275 6183694 89 + 6264404 GCGCGCCGCGAAAAACUUUUCCACGGCACCCUCCCCGGGCCCUCCGGACCUGGUCGCAAGGCCCGCCAUACGGGCCUCGCAACCGUUCA (.((((((.((((....))))..)))).......((((.....))))....(((.((.(((((((.....))))))).)).))))).). ( -35.60) >pau.3414 6456605 87 + 6537648 GCGCGCCGCGAAAAACUUUUCCACGGCACCCUCCCCGGGCACUCCGGACCUGCUCGCAAGGCCCGCCAUACGGGCCUCGCAACCGUU-- (.((((((.((((....))))..)))).......((((.....))))....)).)((.(((((((.....))))))).)).......-- ( -33.60) >pfo.30 60939 52 + 6438405 AUUCUUUUCAAAAAACUUUGG-A----AUAAGCUCAAGAACAAUCCGACCUAAGCCC-------------------------------- .....(((((((....)))))-)----).............................-------------------------------- ( -2.80) >pf5.51 6993214 53 - 7074893 GCCGGUUGCAAAAACUAUUGGCA----AUAAUCGAUGAAAUUUUCGGUGCUAAUAGC-------------------------------- ((.....)).....(((((((((----....((((........))))))))))))).-------------------------------- ( -12.40) >consensus GCGCGCCGCAAAAAACUUUGCCA____ACAAUCCCCGGAACCUCCGGACCUAAUCGC________________________________ ..................................((((.....))))........((.(((((((.....))))))).))......... ( -4.19 = -6.25 + 2.06)

| Location | 6,183,694 – 6,183,783 |
|---|---|
| Length | 89 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 43.32 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -9.91 |
| Energy contribution | -15.97 |
| Covariance contribution | 6.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978755 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.3275 6183694 89 - 6264404 UGAACGGUUGCGAGGCCCGUAUGGCGGGCCUUGCGACCAGGUCCGGAGGGCCCGGGGAGGGUGCCGUGGAAAAGUUUUUCGCGGCGCGC .....((((((((((((((.....))))))))))))))...(((((.....)))))..(.(((((((((((.....))))))))))).) ( -54.80) >pau.3414 6456605 87 - 6537648 --AACGGUUGCGAGGCCCGUAUGGCGGGCCUUGCGAGCAGGUCCGGAGUGCCCGGGGAGGGUGCCGUGGAAAAGUUUUUCGCGGCGCGC --...(.((((((((((((.....)))))))))))).)...(((((.....)))))..(.(((((((((((.....))))))))))).) ( -50.30) >pfo.30 60939 52 - 6438405 --------------------------------GGGCUUAGGUCGGAUUGUUCUUGAGCUUAU----U-CCAAAGUUUUUUGAAAAGAAU --------------------------------(((((((((..........)))))))))..----.-........((((....)))). ( -8.70) >pf5.51 6993214 53 + 7074893 --------------------------------GCUAUUAGCACCGAAAAUUUCAUCGAUUAU----UGCCAAUAGUUUUUGCAACCGGC --------------------------------((((((.(((.(((........))).....----))).))))))............. ( -6.70) >consensus ________________________________GCGAUCAGGUCCGAAGGGCCCGGGGAGGAU____UGCAAAAGUUUUUCGCAACGCGC .........((((((((((.....)))))))))).......(((((.....)))))....(((((((((((.....))))))))))).. ( -9.91 = -15.97 + 6.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:22 2007