
| Sequence ID | pao.2802 |
|---|---|
| Location | 5,308,425 – 5,308,568 |
| Length | 143 |
| Max. P | 0.998592 |

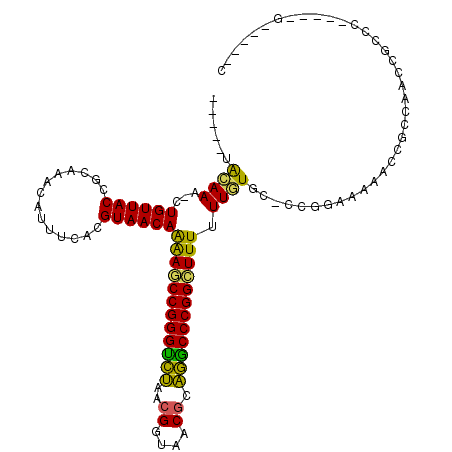
| Location | 5,308,425 – 5,308,528 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.37 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -24.70 |
| Energy contribution | -22.79 |
| Covariance contribution | -1.91 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
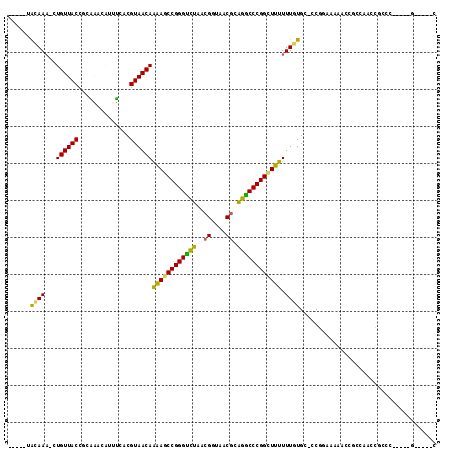
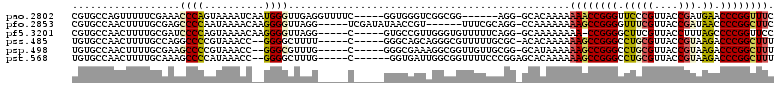
>pao.2802 5308425 103 + 6264404 CACCCUGCAAC-CUGUUACCGCGGACGUACCGCGUAACAGAAACCGGGUUCAUCGGUAACGGGAACCCGGUUUUUUUGUGC-CCU------CCGCCGACCCACC-----GAAAACC ......(((..-(((((((.((((.....)))))))))))(((((((((((.(((....)))))))))))))).....)))-...------.............-----....... ( -38.40) >pfo.2853 5425069 98 + 6438405 -----AAAAAA-CUGUUACCUCAGUCUUUUCACGUAACAGAAGCCGGGGUUAUCGGUAACGAAACCCCGGCUUUUUUUUGG-CCUGCGAAA------ACGGUUAUAUCGA-----C -----....((-(((((..(.(((.((........((.(((((((((((((.(((....)))))))))))))))).)).))-.))).)..)------)))))).......-----. ( -33.80) >pf5.3201 6040802 99 + 7074893 -----AAAAAAACUGUUACCUGCUCCAUCGCACGUAACAGGAACCGGGGCUAAAGGUAACGAAGCCCCGG-UUUUUUUUGC-CCUGAAAAACACCCAACGGCAC-----G-----C -----.......(((((((.(((......))).)))))))(((((((((((...(....)..))))))))-)))....(((-(.((.........))..)))).-----.-----. ( -36.60) >pss.485 5152418 99 - 6093698 -----GACAAA-CUGUUACCUCAAGCGUUUCACGUAACAAAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUGUGU-GCGCAAAAACGCCCUGCUGCCC-----G-----A -----......-............((((((..(((((((((((((((((((..((....)).))))))))))..))))).)-)))...))))))..........-----.-----. ( -32.90) >psp.498 4903650 99 - 5928787 -----UAGAAA-CUGUUACCGCAAACACUUCACGUAACAAAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUAUGC-CCGCAACAACCGCCUUUCGCCC-----G-----C -----..((((-.((((......)))).....(((((.(((((((((((((..((....)).)))))))))))))))))).-..............))))....-----.-----. ( -27.50) >pst.568 5350018 99 - 6397126 -----UGCAAA-CUGUUACCGCAAGCACUUCACGUAACAAAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUGUGCUCCGGGAAAACCGCCAAUCACC------G-----C -----.(((..-..((((((....)........)))))(((((((((((((..((....)).)))))))))))))...)))..(((.....))).........------.-----. ( -31.40) >consensus _____UACAAA_CUGUUACCGCAAACAUUUCACGUAACAAAAGCCGGGUCUAACGGUAACGCAGGCCCGGCUUUUUUGUGC_CCGGAAAAACCGCCAACCGCCC_____G_____C ......((((...((((((..............))))))((((((((((((..((....)).)))))))))))).))))..................................... (-24.70 = -22.79 + -1.91)

| Location | 5,308,425 – 5,308,528 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.37 |
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -19.63 |
| Energy contribution | -19.11 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2802 5308425 103 - 6264404 GGUUUUC-----GGUGGGUCGGCGG------AGG-GCACAAAAAAACCGGGUUCCCGUUACCGAUGAACCCGGUUUCUGUUACGCGGUACGUCCGCGGUAACAG-GUUGCAGGGUG .((((((-----.((......)).)------)))-))......(((((((((((.((....))..)))))))))))(((((((((((.....)))).)))))))-........... ( -40.10) >pfo.2853 5425069 98 - 6438405 G-----UCGAUAUAACCGU------UUUCGCAGG-CCAAAAAAAAGCCGGGGUUUCGUUACCGAUAACCCCGGCUUCUGUUACGUGAAAAGACUGAGGUAACAG-UUUUUU----- .-----.........((((------....(((((-.........(((((((((((((....))).))))))))))))))).))).)(((((((((......)))-))))))----- ( -32.20) >pf5.3201 6040802 99 - 7074893 G-----C-----GUGCCGUUGGGUGUUUUUCAGG-GCAAAAAAAA-CCGGGGCUUCGUUACCUUUAGCCCCGGUUCCUGUUACGUGCGAUGGAGCAGGUAACAGUUUUUUU----- .-----.-----.((((.(((((.....))))))-))).....((-((((((((...........)))))))))).(((((((.(((......))).))))))).......----- ( -41.10) >pss.485 5152418 99 + 6093698 U-----C-----GGGCAGCAGGGCGUUUUUGCGC-ACACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUUUGUUACGUGAAACGCUUGAGGUAACAG-UUUGUC----- (-----(-----((((......((((....))))-.(((..(((((((((((.((((....)))).).)))))))))).....)))....))))))........-......----- ( -35.60) >psp.498 4903650 99 + 5928787 G-----C-----GGGCGAAAGGCGGUUGUUGCGG-GCAUAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUUUGUUACGUGAAGUGUUUGCGGUAACAG-UUUCUA----- .-----.-----.(.(....).).((((..((((-((((..(((((((((((.((((....)))).).))))))))))..........))))))))..))))..-......----- ( -36.90) >pst.568 5350018 99 + 6397126 G-----C------GGUGAUUGGCGGUUUUCCCGGAGCACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUUUGUUACGUGAAGUGCUUGCGGUAACAG-UUUGCA----- (-----(------(..(((((.(((.....)))((((((..(((((((((((.((((....)))).).))))))))))..........)))))).......)))-))))).----- ( -34.60) >consensus G_____C_____GGGCGGUAGGCGGUUUUUCAGG_GCACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUCUGUUACGUGAAAUGCUUGAGGUAACAG_UUUGUA_____ ...........................................((((((((.(((((....))).)).))))))))(((((((..............)))))))............ (-19.63 = -19.11 + -0.52)
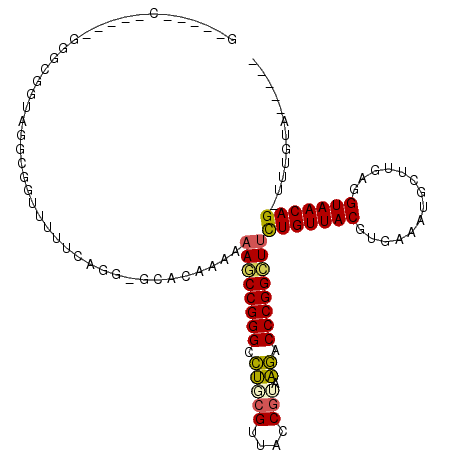
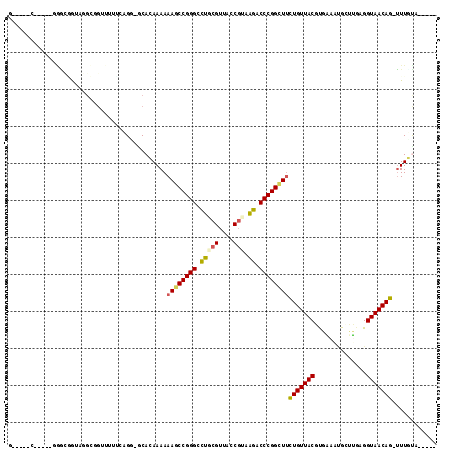
| Location | 5,308,463 – 5,308,568 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.18 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -24.43 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2802 5308463 105 + 6264404 GAAACCGGGUUCAUCGGUAACGGGAACCCGGUUUUUUUGUGC-CCU------CCGCCGACCCACC-----GAAAACCUCAACCCAUUGAUUUUACUGGGUUUCGAAAAACUGGCACG ((((((((((((.(((....)))))))))))))))...((((-(..------.((..((((((..-----((((...((((....))))))))..)))))).)).......))))). ( -37.80) >pfo.2853 5425102 105 + 6438405 GAAGCCGGGGUUAUCGGUAACGAAACCCCGGCUUUUUUUUGG-CCUGCGAAA------ACGGUUAUAUCGA-----CCUAACCCCUUGUUUUAUUGGGGCUCGCAAAAGUUGGCACG ((((((((((((.(((....)))))))))))))))......(-(((((((..------..((((.....))-----))...((((..........)))).)))))......)))... ( -43.60) >pf5.3201 6040836 105 + 7074893 GGAACCGGGGCUAAAGGUAACGAAGCCCCGG-UUUUUUUUGC-CCUGAAAAACACCCAACGGCAC-----G-----CCUAACCCCUUGUUUUACUGGGGAUCGCAAAAGUUGGCACG ((((((((((((...(....)..))))))))-))))......-............(((((((...-----.-----))...((((..........)))).........))))).... ( -35.60) >pss.485 5152451 104 - 6093698 AAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUGUGU-GCGCAAAAACGCCCUGCUGCCC-----G-----AAAAGCCCC--GGUUUACGGGGCCUGGCAAAAGUUGGCACA .(((((((((((..((....)).)))))))))))((((((..-..))))))..(((((..((((.-----(-----....(((((--(.....))))))).))))..))..)))... ( -47.70) >psp.498 4903683 104 - 5928787 AAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUAUGC-CCGCAACAACCGCCUUUCGCCC-----G-----CAAACGCCC--GGUUUACGGGGCUUCGCAAAAGUUGGCACA ((((((((((((..((....)).))))))))))))....(((-(.((.......((.....))..-----(-----(.((.((((--.(....).)))))).))....)).)))).. ( -40.00) >pst.568 5350051 104 - 6397126 AAAGCCGGGUCUUACGGUAACGCAGGCCCGGCUUUUUUGUGCUCCGGGAAAACCGCCAAUCACC------G-----CAAAGCCCC--GGUUUAUGGGGCUUUGCAAAAGUUGGCACA ((((((((((((..((....)).))))))))))))..(((((..(((.....))).((((....------(-----(((((((((--(.....)))))))))))....))))))))) ( -52.90) >consensus AAAGCCGGGUCUAACGGUAACGCAGGCCCGGCUUUUUUGUGC_CCGGAAAAACCGCCAACCGCCC_____G_____CAAAACCCC__GGUUUACGGGGCUUCGCAAAAGUUGGCACA ((((((((((((..((....)).))))))))))))..............................................((((..........)))).................. (-24.43 = -22.80 + -1.63)

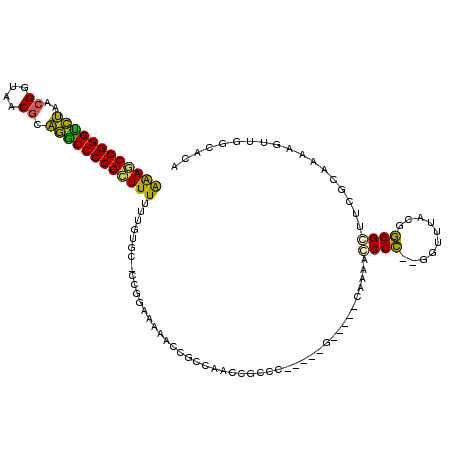
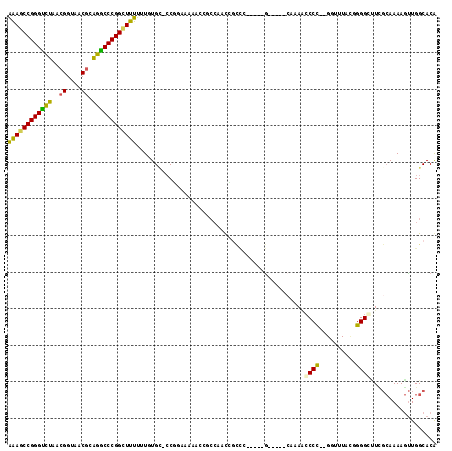
| Location | 5,308,463 – 5,308,568 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.18 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2802 5308463 105 - 6264404 CGUGCCAGUUUUUCGAAACCCAGUAAAAUCAAUGGGUUGAGGUUUUC-----GGUGGGUCGGCGG------AGG-GCACAAAAAAACCGGGUUCCCGUUACCGAUGAACCCGGUUUC .(((((..((..((((..((((.(((((((..........)))))).-----).))))))))..)------).)-))))....(((((((((((.((....))..))))))))))). ( -39.20) >pfo.2853 5425102 105 - 6438405 CGUGCCAACUUUUGCGAGCCCCAAUAAAACAAGGGGUUAGG-----UCGAUAUAACCGU------UUUCGCAGG-CCAAAAAAAAGCCGGGGUUUCGUUACCGAUAACCCCGGCUUC .(.(((......((((((((((..........)))))..((-----(.......)))..------..)))))))-))......((((((((((((((....))).))))))))))). ( -40.80) >pf5.3201 6040836 105 - 7074893 CGUGCCAACUUUUGCGAUCCCCAGUAAAACAAGGGGUUAGG-----C-----GUGCCGUUGGGUGUUUUUCAGG-GCAAAAAAAA-CCGGGGCUUCGUUACCUUUAGCCCCGGUUCC ...(((..(....).((((((..(.....)..)))))).))-----)-----.((((.(((((.....))))))-))).....((-((((((((...........)))))))))).. ( -35.50) >pss.485 5152451 104 + 6093698 UGUGCCAACUUUUGCCAGGCCCCGUAAACC--GGGGCUUUU-----C-----GGGCAGCAGGGCGUUUUUGCGC-ACACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUU (((((...((.(((((((((((((.....)--)))))))..-----.-----.))))).)).(((....)))))-)))....((((((((((.((((....)))).).))))))))) ( -48.80) >psp.498 4903683 104 + 5928787 UGUGCCAACUUUUGCGAAGCCCCGUAAACC--GGGCGUUUG-----C-----GGGCGAAAGGCGGUUGUUGCGG-GCAUAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUU (((((((((..((((.....(((((((((.--....)))))-----)-----)))(....)))))..)))...)-)))))..((((((((((.((((....)))).).))))))))) ( -46.00) >pst.568 5350051 104 + 6397126 UGUGCCAACUUUUGCAAAGCCCCAUAAACC--GGGGCUUUG-----C------GGUGAUUGGCGGUUUUCCCGGAGCACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUU ((((((((...((((((((((((.......--)))))))))-----)------))...))).(((.....)))..)))))..((((((((((.((((....)))).).))))))))) ( -50.40) >consensus CGUGCCAACUUUUGCGAACCCCCGUAAACC__GGGGUUUUG_____C_____GGGCGGUAGGCGGUUUUUCAGG_GCACAAAAAAGCCGGGCCUGCGUUACCGUAAGACCCGGCUUC ..................((((..........))))...............................................((((((((.(((((....))).)).)))))))). (-18.50 = -18.12 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:06:01 2007