
| Sequence ID | pao.2937 |
|---|---|
| Location | 5,541,575 – 5,541,924 |
| Length | 349 |
| Max. P | 0.999132 |

| Location | 5,541,575 – 5,541,669 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
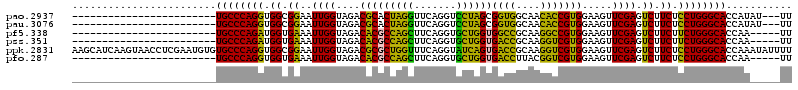
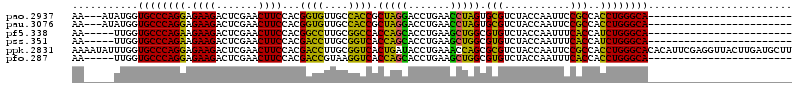
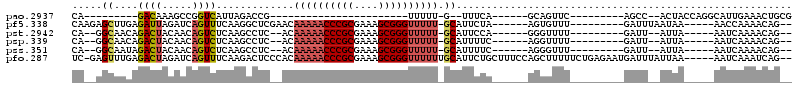
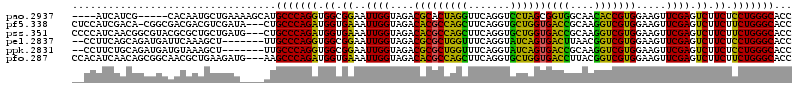
>pao.2937 5541575 94 + 6264404 GUUGUAACACGGAAACGAUGAGCGAUCAUCGACUCCAGUGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACA (((((.((.((....))....(((.(((((((.(((...(((.....)))))).))))).))))).(((((.......)))))..)).))))). ( -30.80) >pf5.338 6403756 67 - 7074893 ------------------------GUCAGCGAUAC---UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGGCCGCAAGG ------------------------((((.((((..---((((.....))))..)))).).))).(((((((.......))))))).((....)) ( -23.30) >pst.2942 5592952 66 + 6397126 -------------------------CUUCCGAUAC---UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGG -------------------------((.(((((..---((((.....))))..))))).))...(((((((.......))))))).((....)) ( -23.20) >psp.339 5255284 66 - 5928787 -------------------------CUUCCGAUAC---UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGG -------------------------((.(((((..---((((.....))))..))))).))...(((((((.......))))))).((....)) ( -23.20) >pss.351 5433975 66 - 6093698 -------------------------CUGUUGAUAC---UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGG -------------------------(((..(((..---((((.....))))..)))..)))...(((((((.......))))))).((....)) ( -21.80) >pfo.287 5820552 66 - 6438405 ------------------------GUUGACGAGA----UGCCCAGGUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCUUACGG ------------------------(((..(((..----((((.....))))...)))...))).(((((((.......)))))))......... ( -17.70) >consensus _________________________CUACCGAUAC___UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGG .........................((((((((.....((((.....))))..))))))))...(((((((.......))))))).((....)) (-17.57 = -17.22 + -0.35)

| Location | 5,541,613 – 5,541,706 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -41.69 |
| Energy contribution | -38.17 |
| Covariance contribution | -3.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.13 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541613 93 + 6264404 ------------------------UGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACACCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAUAU---UU ------------------------((((((((.((.(((.((((....(((.(((((.......))))).((((....))))))).....)))).))).))))))))))......---.. ( -39.00) >pau.3076 5813002 93 + 6537648 ------------------------UGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACACCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAUAU---UU ------------------------((((((((.((.(((.((((....(((.(((((.......))))).((((....))))))).....)))).))).))))))))))......---.. ( -39.00) >pf5.338 6403767 91 - 7074893 ------------------------UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGGCCGCAAGGCCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA-----UU ------------------------((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).))))))))....-----.. ( -42.60) >pss.351 5433985 91 - 6093698 ------------------------UGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA-----UU ------------------------((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).))))))))....-----.. ( -40.50) >ppk.2831 5549297 120 + 6181863 AAGCAUCAAGUAACCUCGAAUGUGUGCCCAGGUGGCGGAAUUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAAAUAUUUU ....................((.(((((((((.((.(((.((((....(((((((((.......))))))((((....))))))).....)))).))).)))))))))))))........ ( -43.40) >pfo.287 5820562 91 - 6438405 ------------------------UGCCCAGGUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCUUACGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAA-----UU ------------------------((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).))))))))....-----.. ( -39.00) >consensus ________________________UGCCCAGGUGGCGAAAUUGGUAGACACGCCAGCUUCAGGUGCUAGUGACCGCAAGGCCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAA_____UU ........................((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).))))))))........... (-41.69 = -38.17 + -3.52)
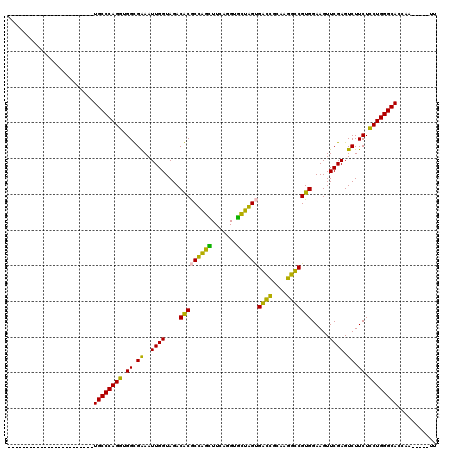
| Location | 5,541,613 – 5,541,706 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -32.33 |
| Energy contribution | -28.78 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541613 93 - 6264404 AA---AUAUGGUGCCCAGGAGAAGACUCGAACUUCCACGGUGUUGCCACCGCUAGGACCUGAACCUAGUGCGUCUACCAAUUCCGCCACCUGGGCA------------------------ ..---......((((((((.((((.......))))..(((.((((..(((((((((.......))))))).))....)))).)))...))))))))------------------------ ( -33.70) >pau.3076 5813002 93 - 6537648 AA---AUAUGGUGCCCAGGAGAAGACUCGAACUUCCACGGUGUUGCCACCGCUAGGACCUGAACCUAGUGCGUCUACCAAUUCCGCCACCUGGGCA------------------------ ..---......((((((((.((((.......))))..(((.((((..(((((((((.......))))))).))....)))).)))...))))))))------------------------ ( -33.70) >pf5.338 6403767 91 + 7074893 AA-----UUGGUGCCCAGAAGAAGACUCGAACUUCCACGGCCUUGCGGCCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCA------------------------ ..-----....((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................))))))))------------------------ ( -33.80) >pss.351 5433985 91 + 6093698 AA-----UUGGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCA------------------------ ..-----....((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................))))))))------------------------ ( -31.70) >ppk.2831 5549297 120 - 6181863 AAAAUAUUUGGUGCCCAGGAGAAGACUCGAACUUCCACGACCUUGCGGUCACUGAUACCUGAAACCAGCGCGUCUACCAAUUCCGCCACCUGGGCACACAUUCGAGGUUACUUGAUGCUU ........(((((((((((.((((.......)))).........((((.....((..((((....))).)..))........))))..))))))))).)).(((((....)))))..... ( -36.12) >pfo.287 5820562 91 + 6438405 AA-----UUGGUGCCCAGGAGAAGACUCGAACUUCCACGACCGUAAGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCACCUGGGCA------------------------ ..-----....((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................))))))))------------------------ ( -36.40) >consensus AA_____UUGGUGCCCAGGAGAAGACUCGAACUUCCACGACCUUGCGGCCACCAGCACCUGAAGCUAGCGCGUCUACCAAUUCCACCACCUGGGCA________________________ ...........((((((((.((((.......))))...((((....)))).(((((.......))))).(((...........)))..))))))))........................ (-32.33 = -28.78 + -3.55)
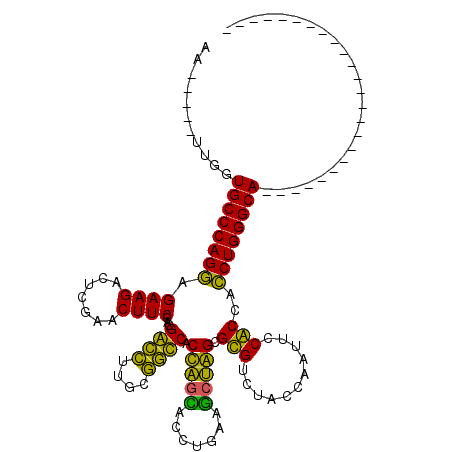
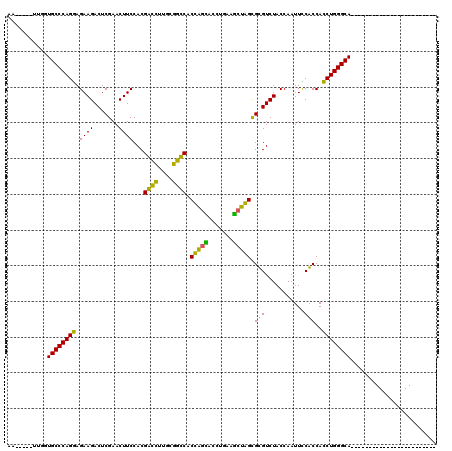
| Location | 5,541,629 – 5,541,730 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -30.21 |
| Energy contribution | -27.55 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541629 101 + 6264404 UUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACACCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAUAU---UUCA---------GACAAAGCCGGUCAUUAGACCG------- .((((...(......)(((((((.....((((((....))))))(((((.......))))))))))))))))...---....---------........(((((....)))))------- ( -33.90) >pf5.338 6403783 115 - 7074893 UUGGUAGACACGCCAGCUUCAGGUGCUGGUGGCCGCAAGGCCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA-----UUCAAGAGCUUGAGAUUAGAUCAGUUUCAAGGCUCGAACAAA ........(((((((((.......))))))((((....)))))))...(((((((((((...((((.......-----(((((....)))))......))))....)))))))))))... ( -47.12) >psp.339 5255310 111 - 5928787 UUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA-----UUCA--GGCAACAGACUACAACAGUCUCAAGCCUC--ACAAA (((((...(((((((((.......))))))((((....))))))).........((((......)))))))))-----...(--(((...(((((.....)))))...)))).--..... ( -40.10) >pss.351 5434001 111 - 6093698 UUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA-----UUCA--GGCAAUAGACUACAACAGUCUCAAGCCUC--ACAAA (((((...(((((((((.......))))))((((....))))))).........((((......)))))))))-----...(--(((...(((((.....)))))...)))).--..... ( -40.10) >ppk.2831 5549337 103 + 6181863 UUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAAAUAUUUUCA--AGUAACAGAUGAC--------CAAGGAUC------- (((((........((((((((((...((.(((((....)))))))((((.......)))).)))))).))))..((((....--)))).......))--------))).....------- ( -26.70) >pfo.287 5820578 114 - 6438405 UUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCUUACGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAA-----UUUC-GAGUUUGAGACUAGAUCAGUUUCAAGACUCCCACAAA .(((....(((((((((.......))))))((((....)))))))((((((.(.((((......)))).).))-----))))-((((((((((((.....)))))).))))))))).... ( -39.60) >consensus UUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAA_____UUCA__AGCAACAGACUACAACAGUCUCAAGACUC__ACAAA .((((...(((((((((.......))))))((((....)))))))...(((((((....)))..))))))))................................................ (-30.21 = -27.55 + -2.66)

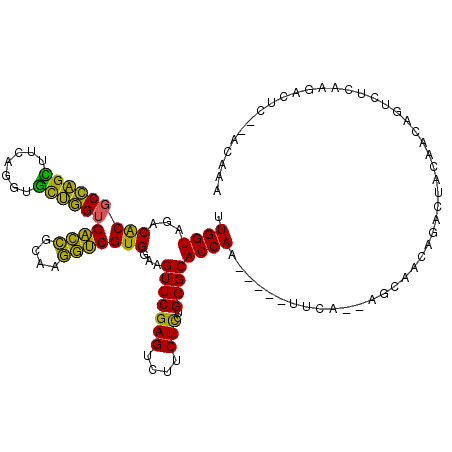
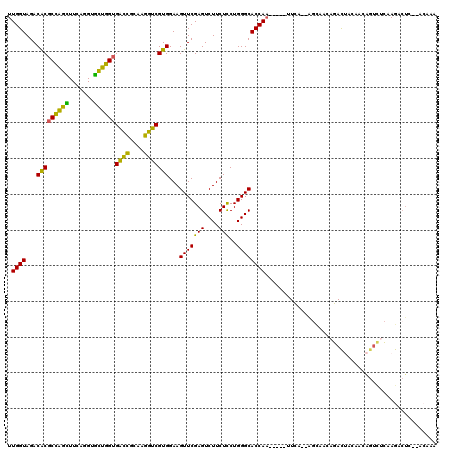
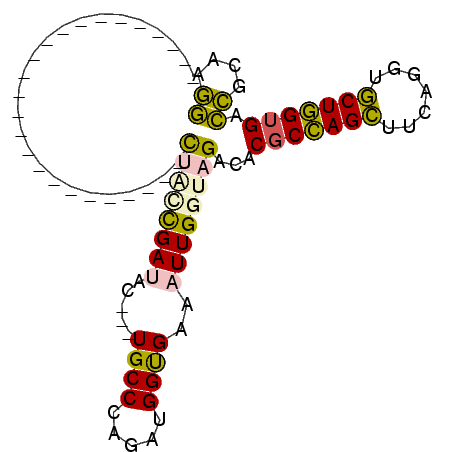
| Location | 5,541,669 – 5,541,745 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -37.19 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541669 76 + 6264404 CCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAUAUUUCA---------GACAAAGCCGGUCAUUAGACCG-----------------------UUUUU-G--UUUCA------GCAG ..((((..(((((((....)))..)))).)))).....(---------(((((((.(((((....)))))-----------------------.))))-)--)))..------.... ( -20.70) >pf5.338 6403823 108 - 7074893 CCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA--UUCAAGAGCUUGAGAUUAGAUCAGUUUCAAGGCUCGAACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUCUA------AGUG ...((((((((((((((((...((((.......--(((((....)))))......))))....)))))))))))((((((((((....))))))))))-...)))))------.... ( -43.52) >pst.2942 5593018 104 + 6397126 UCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA--UUCA--GGCAACAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUCCA------GGGU ((.(((((......((((......)))).....--...(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-)..)))))------.)). ( -40.60) >psp.339 5255350 104 - 5928787 UCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA--UUCA--GGCAACAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGU .(.(((((((....((((......)))).....--...(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-).))))))------).). ( -38.10) >pss.351 5434041 104 - 6093698 UCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA--UUCA--GGCAAUAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGG .(.(((((((....((((......)))).....--...(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-).))))))------).). ( -38.80) >pfo.287 5820618 114 - 6438405 UCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAA--UUUC-GAGUUUGAGACUAGAUCAGUUUCAAGACUCCCACAAAAACCCGCGAAAGCGGGUUUUUUGCAUUCUGCUUUCCAGCU ..(((((((((.(.((((......)))).).))--))))-((((((((((((.....)))))).)))))).)))((((((((((....))))))))))........(((....))). ( -41.40) >consensus UCGUGGAAGUUCGAGUCUUCUUCUGGGCACCAA__UUCA__GGCAACAGACUACAACAGUCUCAAGCCUC__ACAAAAACCCGCGAAAGCGGGUUUUU_GCAUUCUA______AGGG ....((((......((((......))))..............((....((((.....)))).............((((((((((....)))))))))).)).))))........... (-19.21 = -19.68 + 0.47)

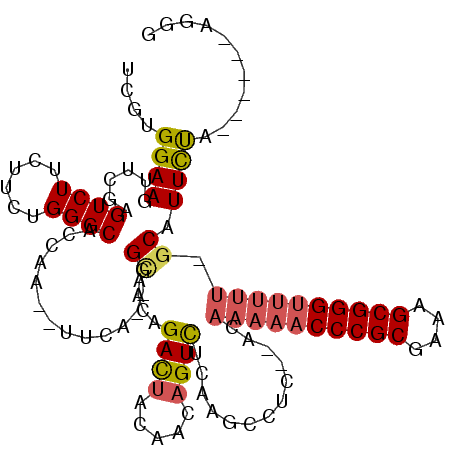
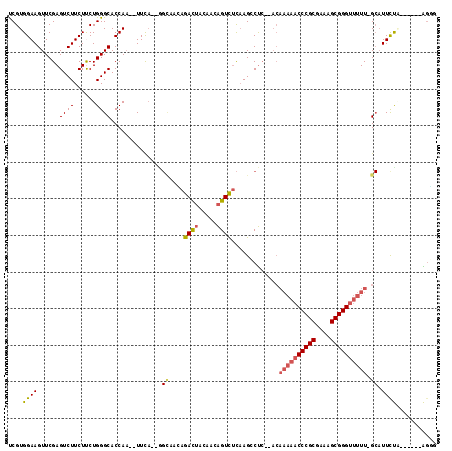
| Location | 5,541,669 – 5,541,745 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -13.50 |
| Energy contribution | -15.57 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541669 76 - 6264404 CUGC------UGAAA--C-AAAAA-----------------------CGGUCUAAUGACCGGCUUUGUC---------UGAAAUAUGGUGCCCAGGAGAAGACUCGAACUUCCACGG ...(------((..(--(-(((..-----------------------(((((....)))))..))))).---------.......(((.......(((....)))......)))))) ( -16.52) >pf5.338 6403823 108 + 7074893 CACU------UAGAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGUUCGAGCCUUGAAACUGAUCUAAUCUCAAGCUCUUGAA--UUGGUGCCCAGAAGAAGACUCGAACUUCCACGG ....------......((-((((((((((....))))))))))))(((((.(((....(((..(((((.((((....)))))--))))....)))...))).))))).......... ( -32.50) >pst.2942 5593018 104 - 6397126 ACCC------UGGAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUGUUGCC--UGAA--UUGGUGCCCAGAAGAAGACUCGAACUUCCACGA ...(------(((...((-((((((((((....))))))))))))--.((((...((((((...))))))...)))--)...--.......))))..((((.......))))..... ( -37.30) >psp.339 5255350 104 + 5928787 ACCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUGUUGCC--UGAA--UUGGUGCCCAGAAGAAGACUCGAACUUCCACGA ....------......((-((((((((((....))))))))))))--((((.(((((.((.......((((..(((--....--..)))...))))...)).))))).))))..... ( -34.60) >pss.351 5434041 104 + 6093698 CCCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUAUUGCC--UGAA--UUGGUGCCCAGAAGAAGACUCGAACUUCCACGA ..((------(.....((-((((((((((....))))))))))))--.((((...((((((...))))))...)))--)...--........)))..((((.......))))..... ( -35.10) >pfo.287 5820618 114 + 6438405 AGCUGGAAAGCAGAAUGCAAAAAACCCGCUUUCGCGGGUUUUUGUGGGAGUCUUGAAACUGAUCUAGUCUCAAACUC-GAAA--UUGGUGCCCAGGAGAAGACUCGAACUUCCACGA .(((....)))........((((((((((....))))))))))((((((((.((((..((..(((.((..(((....-....--)))..))...)))..))..)))))))))))).. ( -38.60) >consensus ACCU______UAAAAUGC_AAAAACCCGCUUUCGCGGGUUUUUGU__GAGGCUUGAGACUGUUGUAGUCUCUUGCC__UGAA__UUGGUGCCCAGAAGAAGACUCGAACUUCCACGA ...................((((((((((....))))))))))....(((.(((.((((((...))))))...............(((...)))....))).)))............ (-13.50 = -15.57 + 2.07)

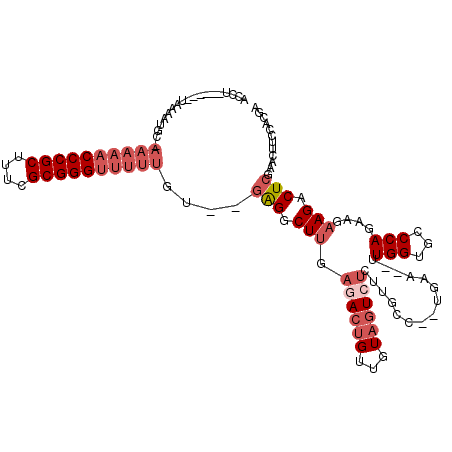
| Location | 5,541,706 – 5,541,774 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.55 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -14.34 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.19 |
| Mean z-score | -6.18 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541706 68 + 6264404 CA---------GACAAAGCCGGUCAUUAGACCG-----------------------UUUUU-G--UUUCA------GCAGUUC---------AGCC--ACUACCAGGCAUUGAAACUGCG .(---------(((((((.(((((....)))))-----------------------.))))-)--)))..------(((((((---------((((--.......)))..)).)))))). ( -25.90) >pf5.338 6403858 97 - 7074893 CAAGAGCUUGAGAUUAGAUCAGUUUCAAGGCUCGAACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUCUA------AGUGUUU---------GAUUUAAUAA-----AACCAAAACAG-- ...((((((((((((.....)))))))).))))((((((((((((((....))))))))))-)..)))..------..(((((---------..........-----.....))))).-- ( -34.56) >pst.2942 5593053 91 + 6397126 CA--GGCAACAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUCCA------GGGUUUU---------GAUU--AUUA-----AAUCAAAACAG-- .(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-).......------..(((((---------((((--....-----)))))))))..-- ( -38.30) >psp.339 5255385 91 - 5928787 CA--GGCAACAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGUUUU---------GAUU--AUUA-----AAUCAAAACAG-- .(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-).......------..(((((---------((((--....-----)))))))))..-- ( -38.30) >pss.351 5434076 91 - 6093698 CA--GGCAAUAGACUACAACAGUCUCAAGCCUC--ACAAAAACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGGUUU---------GAUU--AUUA-----AAUCAAAACAG-- .(--(((...(((((.....)))))...)))).--.(((((((((((....))))))))))-).......------.(..(((---------((((--....-----))))))).)..-- ( -34.20) >pfo.287 5820653 112 - 6438405 UC-GAGUUUGAGACUAGAUCAGUUUCAAGACUCCCACAAAAACCCGCGAAAGCGGGUUUUUUGCAUUCUGCUUUCCAGCUUUUUCUGAGAAUGAUUUAUUAA-----AAUCAAAUCAG-- ..-((((((((((((.....)))))).))))))....((((((((((....))))))))))........(((....))).....((((...((((((....)-----)))))..))))-- ( -36.60) >consensus CA__GGCAACAGACUACAACAGUCUCAAGCCUC__ACAAAAACCCGCGAAAGCGGGUUUUU_GCAUUCUA______AGGGUUU_________GAUU__AUUA_____AAUCAAAACAG__ .....((....((((.....)))).............((((((((((....)))))))))).))........................................................ (-14.34 = -15.28 + 0.95)
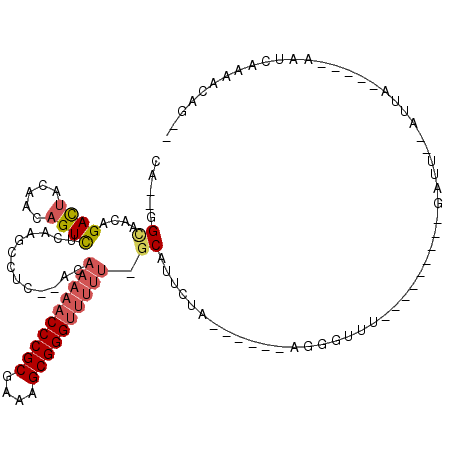
| Location | 5,541,706 – 5,541,774 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.55 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -9.91 |
| Energy contribution | -10.23 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.35 |
| Mean z-score | -4.72 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541706 68 - 6264404 CGCAGUUUCAAUGCCUGGUAGU--GGCU---------GAACUGC------UGAAA--C-AAAAA-----------------------CGGUCUAAUGACCGGCUUUGUC---------UG .(((((((....(((.......--))).---------)))))))------....(--(-(((..-----------------------(((((....)))))..))))).---------.. ( -25.70) >pf5.338 6403858 97 + 7074893 --CUGUUUUGGUU-----UUAUUAAAUC---------AAACACU------UAGAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGUUCGAGCCUUGAAACUGAUCUAAUCUCAAGCUCUUG --.(((((.((((-----(....)))))---------)))))..------..((..((-((((((((((....))))))))))))))((((.((((.............))))))))... ( -30.32) >pst.2942 5593053 91 - 6397126 --CUGUUUUGAUU-----UAAU--AAUC---------AAAACCC------UGGAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUGUUGCC--UG --..(((((((((-----....--))))---------)))))..------......((-((((((((((....))))))))))))--.((((...((((((...))))))...)))--). ( -37.40) >psp.339 5255385 91 + 5928787 --CUGUUUUGAUU-----UAAU--AAUC---------AAAACCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUGUUGCC--UG --..(((((((((-----....--))))---------)))))..------......((-((((((((((....))))))))))))--.((((...((((((...))))))...)))--). ( -37.40) >pss.351 5434076 91 + 6093698 --CUGUUUUGAUU-----UAAU--AAUC---------AAACCCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUUUUUGU--GAGGCUUGAGACUGUUGUAGUCUAUUGCC--UG --....(((((((-----....--))))---------)))....------......((-((((((((((....))))))))))))--.((((...((((((...))))))...)))--). ( -34.80) >pfo.287 5820653 112 + 6438405 --CUGAUUUGAUU-----UUAAUAAAUCAUUCUCAGAAAAAGCUGGAAAGCAGAAUGCAAAAAACCCGCUUUCGCGGGUUUUUGUGGGAGUCUUGAAACUGAUCUAGUCUCAAACUC-GA --((((..(((((-----(....))))))...)))).....(((....))).....(((((((.(((((....))))))))))))..((((.((((.((((...)))).))))))))-.. ( -35.50) >consensus __CUGUUUUGAUU_____UAAU__AAUC_________AAAACCU______UAAAAUGC_AAAAACCCGCUUUCGCGGGUUUUUGU__GAGGCUUGAGACUGUUGUAGUCUCUUGCC__UG .........((((..............................................((((((((((....))))))))))......(((........)))..))))........... ( -9.91 = -10.23 + 0.32)
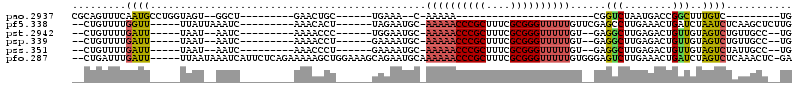
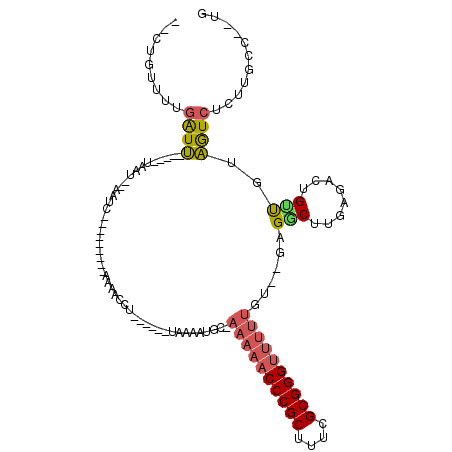
| Location | 5,541,730 – 5,541,805 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -10.69 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
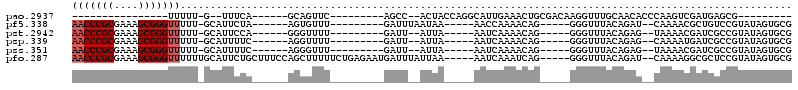
>pao.2937 5541730 75 + 6264404 ----------------UUUUU-G--UUUCA------GCAGUUC---------AGCC--ACUACCAGGCAUUGAAACUGCGACAAGGUUUGCAACACCCAAGUCGAUGAGCG--------- ----------------.....-(--((.((------(((((((---------((((--.......)))..)).))))))(((..(((.......)))...)))..))))).--------- ( -17.80) >pf5.338 6403898 92 - 7074893 AACCCGCGAAAGCGGGUUUUU-GCAUUCUA------AGUGUUU---------GAUUUAAUAA-----AACCAAAACAG-----GGGUUUACAGAU--CAAAACGCUGUCCGUAUAGUGCG (((((((....)))))))...-(((((.((------(((((((---------(((((...((-----..((......)-----)..))...))))--).))))))).....)).))))). ( -30.90) >pst.2942 5593089 90 + 6397126 AACCCGCGAAAGCGGGUUUUU-GCAUUCCA------GGGUUUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCG (((((((....)))))))...-(((((((.------..(((((---------((((--....-----)))))))))..-----))..........--....(((.....)))..))))). ( -31.00) >psp.339 5255421 90 - 5928787 AACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGUUUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--CAAAAUGAUCGCCGUAUAGUGCG (((((((....)))))))...-(((((..(------..(((((---------((((--....-----))))))))).(-----(((((.......--......)))).)))...))))). ( -30.62) >pss.351 5434112 90 - 6093698 AACCCGCGAAAGCGGGUUUUU-GCAUUUUC------AGGGUUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCG (((((((....)))))))...-(((((...------.((.(((---------((((--....-----)))))))...(-----(..(((((...)--))))....)).))....))))). ( -27.40) >pfo.287 5820692 108 - 6438405 AACCCGCGAAAGCGGGUUUUUUGCAUUCUGCUUUCCAGCUUUUUCUGAGAAUGAUUUAUUAA-----AAUCAAAUCAG-----GGGUUUACAGAU--CAAAAGGCGCUCCGUAUAGUGCG (((((((....)))))))....(((((.(((......(((((((((((...((((((....)-----)))))..))))-----.((((....)))--)))))))).....))).))))). ( -36.40) >consensus AACCCGCGAAAGCGGGUUUUU_GCAUUCUA______AGGGUUU_________GAUU__AUUA_____AAUCAAAACAG_____GGGUUUACAGAG__CAAAACGAUCGCCGUAUAGUGCG (((((((....)))))))...................................................................................................... (-10.69 = -11.23 + 0.54)
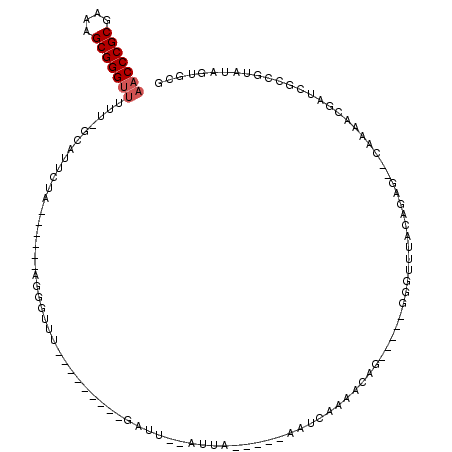
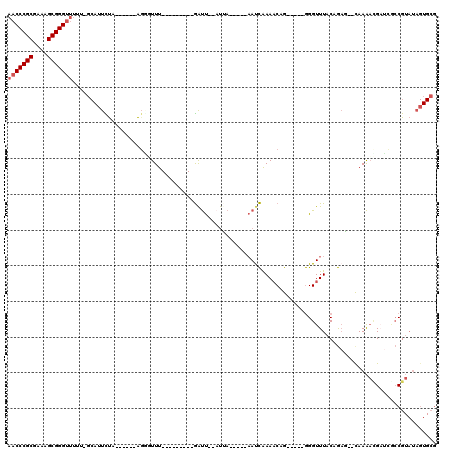
| Location | 5,541,730 – 5,541,805 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -7.69 |
| Energy contribution | -8.23 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541730 75 - 6264404 ---------CGCUCAUCGACUUGGGUGUUGCAAACCUUGUCGCAGUUUCAAUGCCUGGUAGU--GGCU---------GAACUGC------UGAAA--C-AAAAA---------------- ---------...(((..(((..((((.......)))).)))(((((((....(((.......--))).---------)))))))------)))..--.-.....---------------- ( -21.20) >pf5.338 6403898 92 + 7074893 CGCACUAUACGGACAGCGUUUUG--AUCUGUAAACCC-----CUGUUUUGGUU-----UUAUUAAAUC---------AAACACU------UAGAAUGC-AAAAACCCGCUUUCGCGGGUU .(((((((((((((((....)))--.)))))).....-----.(((((.((((-----(....)))))---------)))))..------)))..)))-...(((((((....))))))) ( -24.50) >pst.2942 5593089 90 - 6397126 CGCACUAUACGGCGAUCGUUUUA--CUCUGUAAACCC-----CUGUUUUGAUU-----UAAU--AAUC---------AAAACCC------UGGAAUGC-AAAAACCCGCUUUCGCGGGUU .(((...(((((.(.........--).)))))..((.-----..(((((((((-----....--))))---------)))))..------.))..)))-...(((((((....))))))) ( -26.10) >psp.339 5255421 90 + 5928787 CGCACUAUACGGCGAUCAUUUUG--CUCUGUAAACCC-----CUGUUUUGAUU-----UAAU--AAUC---------AAAACCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUU .(((.((((.(((((.....)))--)).)))).....-----..(((((((((-----....--))))---------)))))..------.....)))-...(((((((....))))))) ( -27.80) >pss.351 5434112 90 + 6093698 CGCACUAUACGGCGAUCGUUUUA--CUCUGUAAACCC-----CUGUUUUGAUU-----UAAU--AAUC---------AAACCCU------GAAAAUGC-AAAAACCCGCUUUCGCGGGUU .(((...(((((.(.........--).))))).....-----....(((((((-----....--))))---------)))....------.....)))-...(((((((....))))))) ( -20.30) >pfo.287 5820692 108 + 6438405 CGCACUAUACGGAGCGCCUUUUG--AUCUGUAAACCC-----CUGAUUUGAUU-----UUAAUAAAUCAUUCUCAGAAAAAGCUGGAAAGCAGAAUGCAAAAAACCCGCUUUCGCGGGUU .(((...((((((.((.....))--.)))))).....-----((((..(((((-----(....))))))...)))).....(((....)))....)))....(((((((....))))))) ( -31.10) >consensus CGCACUAUACGGCGAUCGUUUUG__CUCUGUAAACCC_____CUGUUUUGAUU_____UAAU__AAUC_________AAAACCU______UAAAAUGC_AAAAACCCGCUUUCGCGGGUU ......................................................................................................(((((((....))))))) ( -7.69 = -8.23 + 0.54)
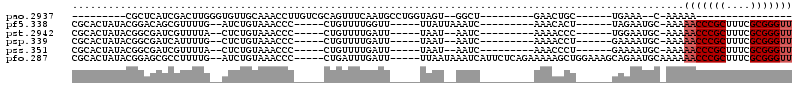
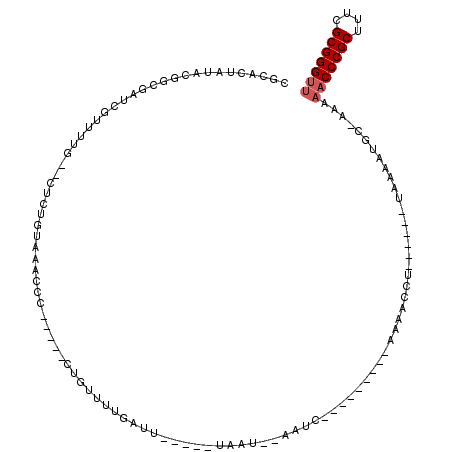
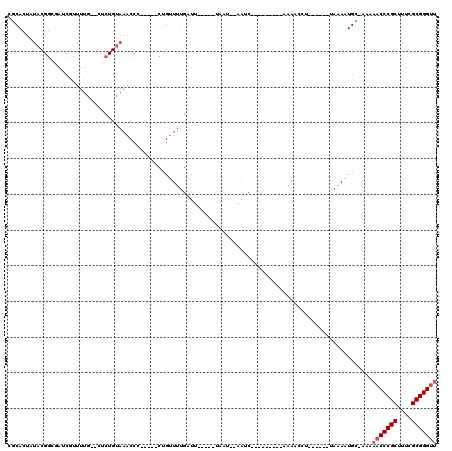
| Location | 5,541,745 – 5,541,836 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.77 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -13.31 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541745 91 + 6264404 UUC---------AGCC--ACUACCAGGCAUUGAAACUGCGACAAGGUUUGCAACACCCAAGUCGAUGAGCG-------------AUCAUCG-----CACAAUGCUGAAAAGCAUGCCCAG ...---------....--.......(((........((((((..(((.......)))...)))(((((...-------------.))))))-----))..(((((....))))))))... ( -23.80) >pf5.338 6403931 95 - 7074893 UUU---------GAUUUAAUAA-----AACCAAAACAG-----GGGUUUACAGAU--CAAAACGCUGUCCGUAUAGUGCGCUCCAUCGACA-CGGCGACGACGUCGAUA---CUGCCCAG (((---------(((((...((-----..((......)-----)..))...))))--)))).((((((....)))))).((...((((((.-((....))..)))))).---..)).... ( -23.50) >pst.2942 5593122 94 + 6397126 UUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCGCCCCAUCGACGGCGGAGACGCUGCUGAUG---CUGCCCAG (((---------((((--....-----)))))))....-----((((.......(--((..(((.....)))....)))....(((((.(((((....))))).)))))---..)))).. ( -27.80) >psp.339 5255454 94 - 5928787 UUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--CAAAAUGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGGAGACGCUGCUGAUG---CUGCCCAG (((---------((((--....-----)))))))....-----((((.......(--((..(((.....)))....)))....(((((.(((((....))))).)))))---..)))).. ( -28.70) >pss.351 5434145 94 - 6093698 UUU---------GAUU--AUUA-----AAUCAAAACAG-----GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGUACGCGCUGCUGAUG---CUGCCCAG (((---------((((--....-----)))))))....-----((((...(((((--(...(((.....)))...(((((((........)))))))..))).)))...---..)))).. ( -27.30) >pfo.287 5820732 105 - 6438405 UUUUCUGAGAAUGAUUUAUUAA-----AAUCAAAUCAG-----GGGUUUACAGAU--CAAAAGGCGCUCCGUAUAGUGCGCCACAUCAACAGCGGCAACGCUGAAGAUG---AAGCCCAG ....((((...((((((....)-----)))))..))))-----((((((......--.....(((((.(......).))))).((((..(((((....)))))..))))---)))))).. ( -42.30) >consensus UUU_________GAUU__AUUA_____AAUCAAAACAG_____GGGUUUACAGAG__CAAAACGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGGAGACGCUGCUGAUG___CUGCCCAG ...........................................((((......................(((.....)))...(((((.(((((....))))).))))).....)))).. (-13.31 = -14.97 + 1.66)

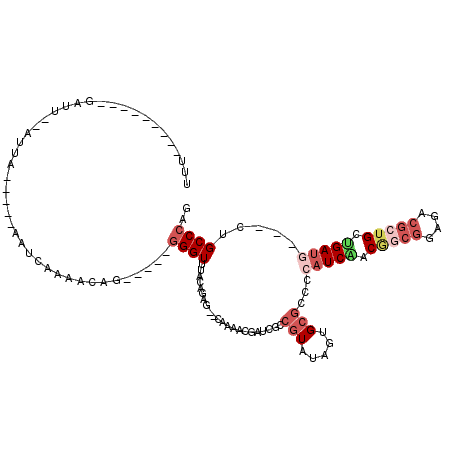

| Location | 5,541,774 – 5,541,876 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -28.47 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541774 102 + 6264404 ACAAGGUUUGCAACACCCAAGUCGAUGAGCG-------------AUCAUCG-----CACAAUGCUGAAAAGCAUGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCG .....((((((.....((..((((((((...-------------.))))))-----.)).(((((....)))))(((.....))))).....))))))...(((((.......))))).. ( -32.20) >pf5.338 6403955 111 - 7074893 ---GGGUUUACAGAU--CAAAACGCUGUCCGUAUAGUGCGCUCCAUCGACA-CGGCGACGACGUCGAUA---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG ---..((((((.(((--.....(((((((.(..((((.......((((((.-((....))..)))))))---)))..).)))))))..))).)))))).(((((((.......))))))) ( -34.81) >pst.2942 5593144 112 + 6397126 ---GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCGCCCCAUCGACGGCGGAGACGCUGCUGAUG---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG ---..((((((...(--.....)..(((((((....((.((..(((((.(((((....))))).)))))---..)).)).))))))).....)))))).(((((((.......))))))) ( -44.20) >psp.339 5255476 112 - 5928787 ---GGGUUUACAGAG--CAAAAUGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGGAGACGCUGCUGAUG---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG ---..((((((....--...(((..(((((((....((.((..(((((.(((((....))))).)))))---..)).)).))))))).))).)))))).(((((((.......))))))) ( -44.70) >pss.351 5434167 112 - 6093698 ---GGGUUUACAGAG--UAAAACGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGUACGCGCUGCUGAUG---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG ---..((((((...(--.....)..(((((((....((.((..(((((.(((((....))))).)))))---..)).)).))))))).....)))))).(((((((.......))))))) ( -41.70) >pfo.287 5820765 112 - 6438405 ---GGGUUUACAGAU--CAAAAGGCGCUCCGUAUAGUGCGCCACAUCAACAGCGGCAACGCUGAAGAUG---AAGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG ---((((((......--.....(((((.(......).))))).((((..(((((....)))))..))))---)))))).....................(((((((.......))))))) ( -46.00) >consensus ___GGGUUUACAGAG__CAAAACGAUCGCCGUAUAGUGCGCCCCAUCAACGGCGGAGACGCUGCUGAUG___CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUG .....((((((................................(((((.(((((....))))).))))).....(((.....))).......)))))).(((((((.......))))))) (-28.47 = -28.92 + 0.45)

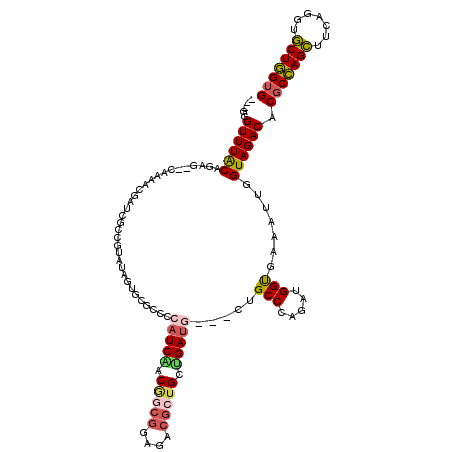
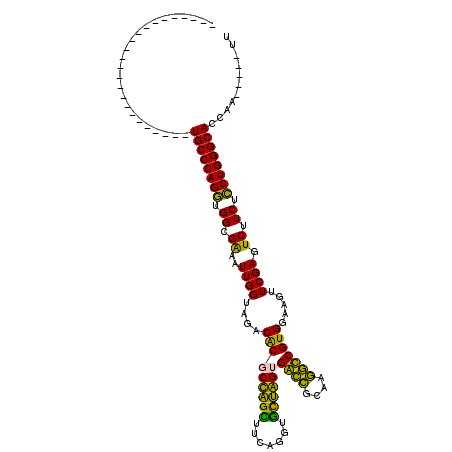
| Location | 5,541,805 – 5,541,916 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -39.77 |
| Energy contribution | -36.22 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
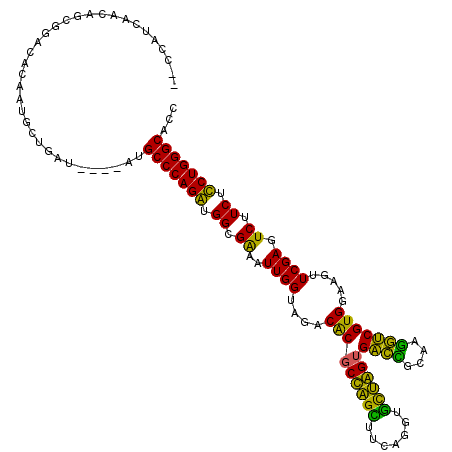
>pao.2937 5541805 111 + 6264404 ----AUCAUCG-----CACAAUGCUGAAAAGCAUGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACACCGUGGAAGUUCGAGUCUUCUCCUGGGCACC ----.......-----....(((((....)))))(((((((.((.(((.((((....(((.(((((.......))))).((((....))))))).....)))).))).)))))))))... ( -45.10) >pf5.338 6403990 116 - 7074893 CUCCAUCGACA-CGGCGACGACGUCGAUA---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACC ....((((((.-((....))..)))))).---.((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))).. ( -50.10) >pss.351 5434202 117 - 6093698 CCCCAUCAACGGCGUACGCGCUGCUGAUG---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACC ...(((((.(((((....))))).)))))---.((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))).. ( -53.80) >pel.2837 5236275 111 + 5888780 --CCUUCAGCAGAUGAUUCAAAGCU-------UUGCCCAGGUGGCGGAAUUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACUUAACGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACC --.....(((.((....))...)))-------.((((((((.((.(((.((((....(((((((((.......))))))((((....))))))).....)))).))).)))))))))).. ( -33.50) >ppk.2831 5549472 111 + 6181863 --CCUUCUGCAGAUGAUGUAAAGCU-------UUGCCCAGGUGGCGGAAUUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACC --.(((.((((.....)))))))..-------.((((((((.((.(((.((((....(((((((((.......))))))((((....))))))).....)))).))).)))))))))).. ( -41.10) >pfo.287 5820800 117 - 6438405 CCACAUCAACAGCGGCAACGCUGAAGAUG---AAGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCUUACGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACC ...((((..(((((....)))))..))))---..(((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))... ( -52.50) >consensus __CCAUCAACAGCGGACACAAUGCUGAU____AUGCCCAGAUGGCGAAAUUGGUAGACACGCCAGCUUCAGGUGCUAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACC ..................................(((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))... (-39.77 = -36.22 + -3.55)
| Location | 5,541,805 – 5,541,916 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -37.89 |
| Consensus MFE | -29.66 |
| Energy contribution | -27.36 |
| Covariance contribution | -2.30 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541805 111 - 6264404 GGUGCCCAGGAGAAGACUCGAACUUCCACGGUGUUGCCACCGCUAGGACCUGAACCUAGUGCGUCUACCAAUUCCGCCACCUGGGCAUGCUUUUCAGCAUUGUG-----CGAUGAU---- .(((((((((.((((.......))))..(((.((((..(((((((((.......))))))).))....)))).)))...)))))))))(((....)))......-----.......---- ( -37.70) >pf5.338 6403990 116 + 7074893 GGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCAG---UAUCGACGUCGUCGCCG-UGUCGAUGGAG ..((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................)))))))).---((((((((.((....))-))))))))... ( -42.40) >pss.351 5434202 117 + 6093698 GGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCAG---CAUCAGCAGCGCGUACGCCGUUGAUGGGG ..((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................)))))))).---(((((((.(((....))).)))))))... ( -45.70) >pel.2837 5236275 111 - 5888780 GGUGCCCAGGAGAAGACUCGAACUUCCACGACCGUUAAGUCACUGAUACCUGAAACCAGCGCGUCUACCAAUUCCGCCACCUGGGCAA-------AGCUUUGAAUCAUCUGCUGAAGG-- ..((((((((.((((.......))))...(((((((...(((........)))....)))).)))..............)))))))).-------(((...((....)).))).....-- ( -27.70) >ppk.2831 5549472 111 - 6181863 GGUGCCCAGGAGAAGACUCGAACUUCCACGACCUUGCGGUCACUGAUACCUGAAACCAGCGCGUCUACCAAUUCCGCCACCUGGGCAA-------AGCUUUACAUCAUCUGCAGAAGG-- ..((((((((.((((.......)))).........((((.....((..((((....))).)..))........))))..)))))))).-------.((............))......-- ( -28.82) >pfo.287 5820800 117 + 6438405 GGUGCCCAGAAGAAGACUCGAACUUCCACGACCGUAAGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCUU---CAUCUUCAGCGUUGCCGCUGUUGAUGUGG ...(((((((.((((.......))))(((((((....)))).(((((.......))))).)))................)))))))..---((((..(((((....)))))..))))... ( -45.00) >consensus GGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUAGCGCGUCUACCAAUUCCACCACCUGGGCAA____AUCAGCAUCGUAUCCGCUGUUGAUGG__ ..((((((((...((((.((.........((((....)))).(((((.......))))))).)))).............))))))))................................. (-29.66 = -27.36 + -2.30)
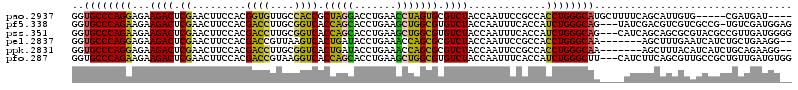
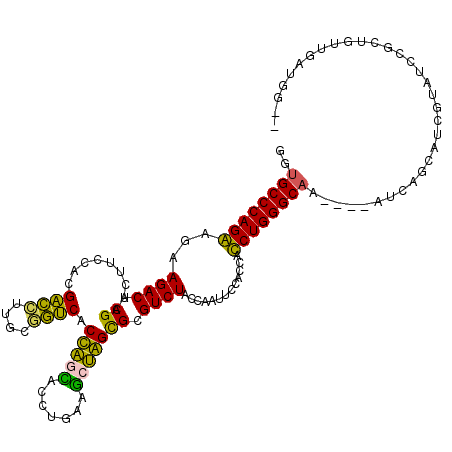
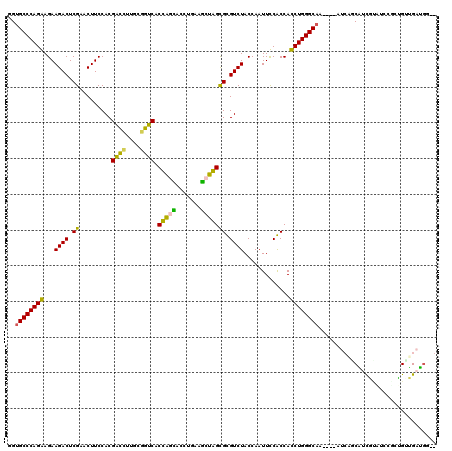
| Location | 5,541,809 – 5,541,924 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -39.77 |
| Energy contribution | -36.22 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541809 115 + 6264404 UCG-----CACAAUGCUGAAAAGCAUGCCCAGGUGGCGGAAUUGGUAGACGCACUAGGUUCAGGUCCUAGCGGUGGCAACACCGUGGAAGUUCGAGUCUUCUCCUGGGCACCAUCUUCAA ...-----....(((((....)))))(((((((.((.(((.((((....(((.(((((.......))))).((((....))))))).....)))).))).)))))))))........... ( -45.10) >pf5.338 6403998 114 - 7074893 ACA-CGGCGACGACGUCGAUA---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCA-AUUCA- ...-(((((....)))))...---.((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).))))))))...-.....- ( -46.50) >pss.351 5434210 115 - 6093698 ACGGCGUACGCGCUGCUGAUG---CUGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCA-AUUCA- .(((((....)))))....((---.((((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))).))-.....- ( -48.90) >pel.2837 5236281 110 + 5888780 GCAGAUGAUUCAAAGCU-------UUGCCCAGGUGGCGGAAUUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACUUAACGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACC--AUACA- .................-------.((((((((.((.(((.((((....(((((((((.......))))))((((....))))))).....)))).))).))))))))))..--.....- ( -32.90) >ppk.2831 5549478 110 + 6181863 GCAGAUGAUGUAAAGCU-------UUGCCCAGGUGGCGGAAUUGGUAGACGCGCUGGUUUCAGGUAUCAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACC--AUACA- (((.....)))......-------.((((((((.((.(((.((((....(((((((((.......))))))((((....))))))).....)))).))).))))))))))..--.....- ( -39.50) >pfo.287 5820808 115 - 6438405 ACAGCGGCAACGCUGAAGAUG---AAGCCCAGAUGGUGAAAUUGGUAGACACGCCAGCUUCAGGUGCUGGUGACCUUACGGUCGUGGAAGUUCGAGUCUUCUUCUGGGCACCA-AUUCA- .(((((....)))))......---..(((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))....-.....- ( -47.40) >consensus ACAGCGGACACAAUGCUGAU____AUGCCCAGAUGGCGAAAUUGGUAGACACGCCAGCUUCAGGUGCUAGUGACCGCAAGGUCGUGGAAGUUCGAGUCUUCUCCUGGGCACCA_AUUCA_ ..........................(((((((.((.((..((((....(((((((((.......))))))((((....))))))).....)))).)).)).)))))))........... (-39.77 = -36.22 + -3.55)
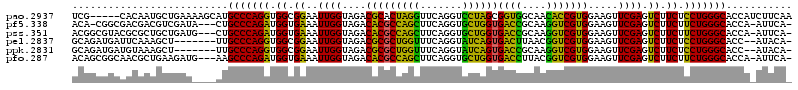
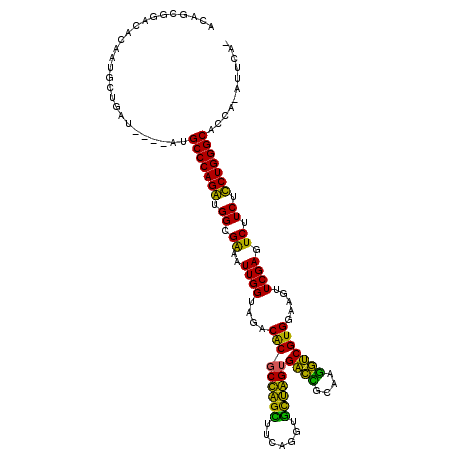
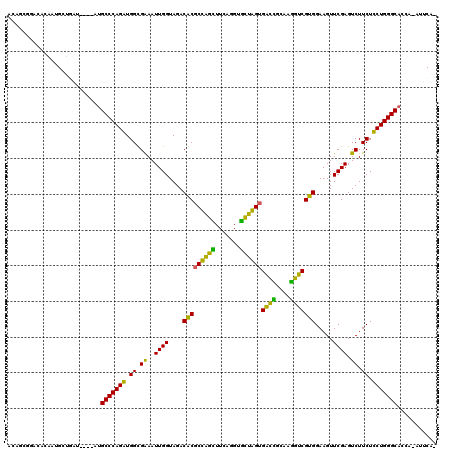
| Location | 5,541,809 – 5,541,924 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -29.66 |
| Energy contribution | -27.36 |
| Covariance contribution | -2.30 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2937 5541809 115 - 6264404 UUGAAGAUGGUGCCCAGGAGAAGACUCGAACUUCCACGGUGUUGCCACCGCUAGGACCUGAACCUAGUGCGUCUACCAAUUCCGCCACCUGGGCAUGCUUUUCAGCAUUGUG-----CGA .((((((..(((((((((.((((.......))))..(((.((((..(((((((((.......))))))).))....)))).)))...)))))))))..))))))((.....)-----).. ( -42.30) >pf5.338 6403998 114 + 7074893 -UGAAU-UGGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCAG---UAUCGACGUCGUCGCCG-UGU -.....-.((((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................)))))))..---....(((...)))))).-... ( -35.90) >pss.351 5434210 115 + 6093698 -UGAAU-UGGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCAG---CAUCAGCAGCGCGUACGCCGU -(((..-((.((((((((.((((.......))))(((((((....)))).(((((.......))))).)))................)))))))).---)))))((.(((....))).)) ( -39.30) >pel.2837 5236281 110 - 5888780 -UGUAU--GGUGCCCAGGAGAAGACUCGAACUUCCACGACCGUUAAGUCACUGAUACCUGAAACCAGCGCGUCUACCAAUUCCGCCACCUGGGCAA-------AGCUUUGAAUCAUCUGC -.....--..((((((((.((((.......))))...(((((((...(((........)))....)))).)))..............)))))))).-------................. ( -25.90) >ppk.2831 5549478 110 - 6181863 -UGUAU--GGUGCCCAGGAGAAGACUCGAACUUCCACGACCUUGCGGUCACUGAUACCUGAAACCAGCGCGUCUACCAAUUCCGCCACCUGGGCAA-------AGCUUUACAUCAUCUGC -((((.--((((((((((.((((.......)))).........((((.....((..((((....))).)..))........))))..)))))))..-------.))).))))........ ( -30.52) >pfo.287 5820808 115 + 6438405 -UGAAU-UGGUGCCCAGAAGAAGACUCGAACUUCCACGACCGUAAGGUCACCAGCACCUGAAGCUGGCGUGUCUACCAAUUUCACCAUCUGGGCUU---CAUCUUCAGCGUUGCCGCUGU -.....-(((.(((((((.((((.......))))(((((((....)))).(((((.......))))).)))................))))))).)---))....(((((....))))). ( -41.60) >consensus _UGAAU_UGGUGCCCAGAAGAAGACUCGAACUUCCACGACCUUGCGGUCACCAGCACCUGAAGCUAGCGCGUCUACCAAUUCCACCACCUGGGCAA____AUCAGCAUCGUAUCCGCUGU ..........((((((((...((((.((.........((((....)))).(((((.......))))))).)))).............))))))))......................... (-29.66 = -27.36 + -2.30)
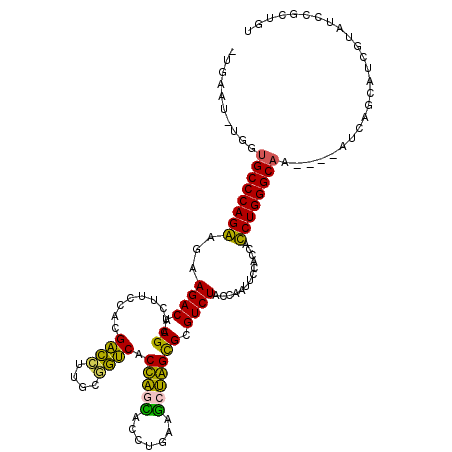
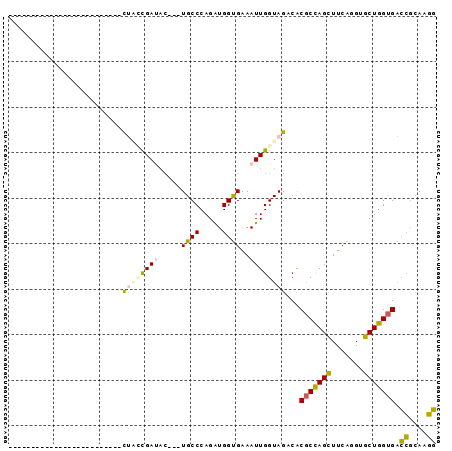
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:52 2007