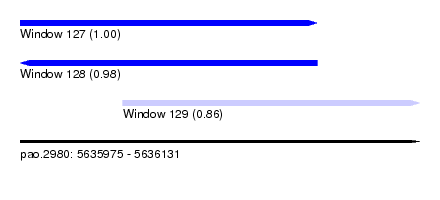
| Sequence ID | pao.2980 |
|---|---|
| Location | 5,635,975 – 5,636,131 |
| Length | 156 |
| Max. P | 0.995955 |

| Location | 5,635,975 – 5,636,091 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.99 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
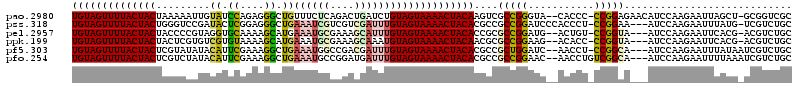
>pao.2980 5635975 116 + 6264404 UGUAGUUUUACUACUAAAAAUUGUAUCCAGAGGGCUGUUUCUCAGACUGAUCUGUAGUAAAACUACAAGUCGCCGGGUA--CACCC-CCGGAGAACAUCCAAGAAUUAGCU-GCGGUCGC ((((((((((((((...............(((((....)))))(((....)))))))))))))))))..((.(((((..--....)-)))).)).(..((.((......))-..))..). ( -32.80) >pss.318 5514594 115 - 6093698 UGUAGUUUUACUACUGGGUCCGAUACUCGGAGGGCUGAAAUCGUCGUCGAUUUGUAGUAAAACUACACGCCGCCGGAUCCCACCCU-CCGGAA---AUCCAAGAAUUUAUG-UCGUCUGC ((((((((((((((..(((((..........))))).((((((....)))))))))))))))))))).....(((((........)-))))..---...............-........ ( -35.60) >pel.2957 5464775 113 + 5888780 UGUAGUUUUACUACUACCCCGUAGGUGCAAAAGCAUGAAAUGCGAAAGCAUUUGUAGUAAAACUACACCGCGCCGGAUG--ACUGU-CCGGUA---AUCCAAGAAUUCACG-ACGUCUGC ((((((((((((((..........((((....)))).((((((....))))))))))))))))))))....(((((((.--...))-))))).---...............-........ ( -41.20) >ppk.199 5771025 113 - 6181863 UGUAGUUUUACUACUACUCGUGUCGUGUAAAAGCAUGAAAUGCGAAAGCAAAUGUAGUAAAACUACAACGCGCCGGAAG--ACACC-CCGGUA---AUCCAAGAAUUCACG-ACGUCUGC ((((((((((((((........((((((....))))))..(((....)))...))))))))))))))....(((((...--.....-))))).---...............-........ ( -37.40) >pf5.303 6480924 114 - 7074893 UGUAGUUUUACUACUCGUAUAUACAUUCGAAAGGCUGAAAUGGCCGACGAUUUGUAGUAAAACUACACGCCGCUGGAUC--AACCU-CCGGCA---AUCCAAGAAUUUAUAAUCGUCUGC ((((((((((((((..((....))..(((...((((.....))))..)))...)))))))))))))).((((..((...--..)).-.)))).---........................ ( -34.80) >pfo.254 5902164 115 - 6438405 UGUAGUUUUACUACUCGUCUAUACAUUCGAAAGGCUGAAAUGCCGGAUGAUUUGUAGUAAAACUACACGCCGCCGGAAC--AACCUGUCGGCA---AUCCAAGAAUUUUAAAUCGUCUGC ((((((((((((((((((((...(....)...(((......)))))))))...)))))))))))))).((((.(((...--...))).)))).---........................ ( -36.60) >consensus UGUAGUUUUACUACUAGUCCAUACAUUCAAAAGGCUGAAAUGCCAAACGAUUUGUAGUAAAACUACACGCCGCCGGAUC__AACCU_CCGGAA___AUCCAAGAAUUUACG_UCGUCUGC ((((((((((((((.........((.((....)).))((((((....))))))))))))))))))))....(((((...........)))))............................ (-24.61 = -24.87 + 0.26)
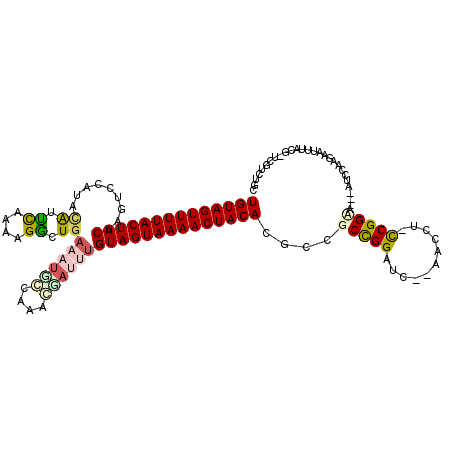
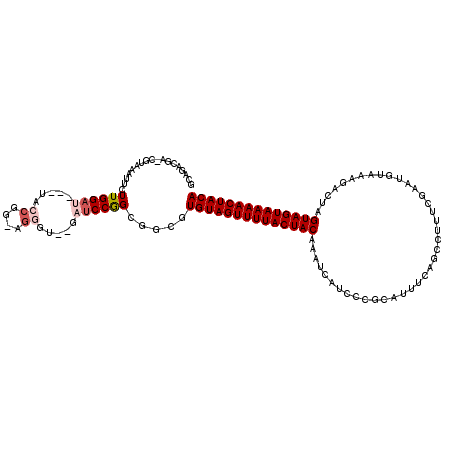
| Location | 5,635,975 – 5,636,091 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.99 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2980 5635975 116 - 6264404 GCGACCGC-AGCUAAUUCUUGGAUGUUCUCCGG-GGGUG--UACCCGGCGACUUGUAGUUUUACUACAGAUCAGUCUGAGAAACAGCCCUCUGGAUACAAUUUUUAGUAGUAAAACUACA .((.(((.-.....(((((((((.....)))))-)))).--....)))))...(((((((((((((((((...(((((((........))).))))......))).)))))))))))))) ( -35.00) >pss.318 5514594 115 + 6093698 GCAGACGA-CAUAAAUUCUUGGAU---UUCCGG-AGGGUGGGAUCCGGCGGCGUGUAGUUUUACUACAAAUCGACGACGAUUUCAGCCCUCCGAGUAUCGGACCCAGUAGUAAAACUACA ........-.........((.(((---...(((-((((((..(((((.(((..(((((.....)))))..))).))..)))..)).)))))))...))).))....((((.....)))). ( -37.80) >pel.2957 5464775 113 - 5888780 GCAGACGU-CGUGAAUUCUUGGAU---UACCGG-ACAGU--CAUCCGGCGCGGUGUAGUUUUACUACAAAUGCUUUCGCAUUUCAUGCUUUUGCACCUACGGGGUAGUAGUAAAACUACA .......(-((((((((....)))---).((((-(....--..))))))))))((((((((((((((((((((....))))))..(((....)))((....))...)))))))))))))) ( -40.20) >ppk.199 5771025 113 + 6181863 GCAGACGU-CGUGAAUUCUUGGAU---UACCGG-GGUGU--CUUCCGGCGCGUUGUAGUUUUACUACAUUUGCUUUCGCAUUUCAUGCUUUUACACGACACGAGUAGUAGUAAAACUACA ...(.(((-((.(((.((((((..---..))))-))...--.)))))))))..((((((((((((((...(((....))).....(((((...........))))))))))))))))))) ( -34.70) >pf5.303 6480924 114 + 7074893 GCAGACGAUUAUAAAUUCUUGGAU---UGCCGG-AGGUU--GAUCCAGCGGCGUGUAGUUUUACUACAAAUCGUCGGCCAUUUCAGCCUUUCGAAUGUAUAUACGAGUAGUAAAACUACA ....((.......((((....)))---)((((.-.((..--...))..))))))(((((((((((((...(((..(((.......)))...))).(((....))).))))))))))))). ( -32.40) >pfo.254 5902164 115 + 6438405 GCAGACGAUUUAAAAUUCUUGGAU---UGCCGACAGGUU--GUUCCGGCGGCGUGUAGUUUUACUACAAAUCAUCCGGCAUUUCAGCCUUUCGAAUGUAUAGACGAGUAGUAAAACUACA ....(((((((((.....))))))---)((((...((..--...))..))))))(((((((((((((.........(((......)))..(((..........)))))))))))))))). ( -34.40) >consensus GCAGACGA_CGUAAAUUCUUGGAU___UACCGG_AGGGU__GAUCCGGCGGCGUGUAGUUUUACUACAAAUCAUCCCGCAUUUCAGCCUUUCGAAUGUAAAGACUAGUAGUAAAACUACA ..................((((((.....((....)).....)))))).....((((((((((((((.......................................)))))))))))))) (-17.55 = -18.41 + 0.86)

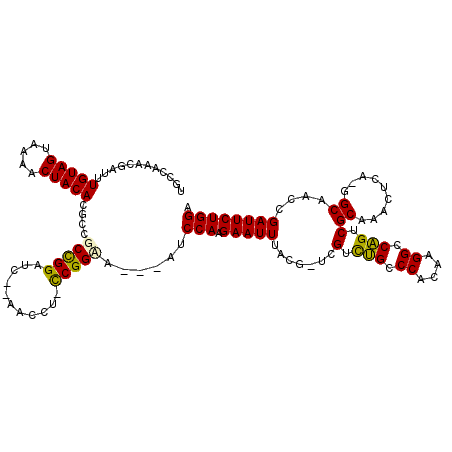
| Location | 5,636,015 – 5,636,131 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -21.27 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2980 5636015 116 + 6264404 CUCAGACUGAUCUGUAGUAAAACUACAAGUCGCCGGGUA--CACCC-CCGGAGAACAUCCAAGAAUUAGCU-GCGGUCGCCCACGAGGCCGGCCGCUCACCUAUGGCAUCCGAUUCUGGA .(((((..((..(((((.....)))))..))((((((..--....)-))(((.....)))......(((..-(((((((.((....)).)))))))....))).))).......))))). ( -40.50) >pss.318 5514634 114 - 6093698 UCGUCGUCGAUUUGUAGUAAAACUACACGCCGCCGGAUCCCACCCU-CCGGAA---AUCCAAGAAUUUAUG-UCGUCUGCCCACAAGGCCAGUCGCAAACUCA-GGCAACCGAUUCUGGA (((..((((...(((((.....))))).(((.(((((........)-))))..---.............((-.((.(((.((....)).))).))))......-)))...))))..))). ( -29.70) >pel.2957 5464815 112 + 5888780 UGCGAAAGCAUUUGUAGUAAAACUACACCGCGCCGGAUG--ACUGU-CCGGUA---AUCCAAGAAUUCACG-ACGUCUGCCCAUAAGGCCAGUCGCAAACUCA-GGCCAUCGAUUCUGGU (((....)))..(((((.....)))))....(((((((.--...))-))))).---..(((.(((((...(-((....(((.....)))..)))((.......-.))....)))))))). ( -32.80) >ppk.199 5771065 112 - 6181863 UGCGAAAGCAAAUGUAGUAAAACUACAACGCGCCGGAAG--ACACC-CCGGUA---AUCCAAGAAUUCACG-ACGUCUGCCCAUAAGGCCAGUCGCAAUCUCA-GGCAAUCGAUUCUGGU (((....)))..(((((.....)))))....(((((...--.....-))))).---..(((.(((((...(-((....(((.....)))..)))((.......-.))....)))))))). ( -31.30) >pf5.303 6480964 113 - 7074893 UGGCCGACGAUUUGUAGUAAAACUACACGCCGCUGGAUC--AACCU-CCGGCA---AUCCAAGAAUUUAUAAUCGUCUGCCCAAAAGGCCAGUCGCAAACUCA-GGCAACCGAUUCUGGA .(((.((((((((((((.....))))).((((..((...--..)).-.)))).---..............))))))).)))......((((((.....)))..-)))..(((....))). ( -32.70) >pfo.254 5902204 114 - 6438405 UGCCGGAUGAUUUGUAGUAAAACUACACGCCGCCGGAAC--AACCUGUCGGCA---AUCCAAGAAUUUUAAAUCGUCUGCCCACAAGGCCAGUCGCAAACUCA-GGCAACCGAUUCUGGA ((((((((....(((((.....))))).((((.(((...--...))).)))).---)))).............((.(((.((....)).))).))........-)))).(((....))). ( -33.40) >consensus UGCCAAACGAUUUGUAGUAAAACUACACGCCGCCGGAUC__AACCU_CCGGAA___AUCCAAGAAUUUACG_UCGUCUGCCCACAAGGCCAGUCGCAAACUCA_GGCAACCGAUUCUGGA ............(((((.....)))))....(((((...........)))))......(((.(((((.......(.(((.((....)).))).)((.........))....)))))))). (-21.27 = -20.80 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:35 2007