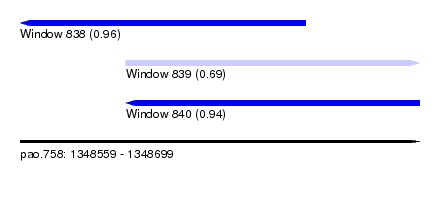
| Sequence ID | pao.758 |
|---|---|
| Location | 1,348,559 – 1,348,699 |
| Length | 140 |
| Max. P | 0.962229 |

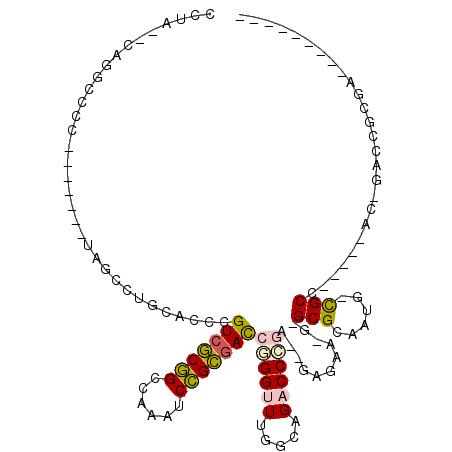
| Location | 1,348,559 – 1,348,659 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 57.69 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.30 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
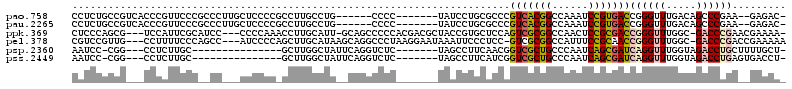
>pao.758 1348559 100 - 6264404 CCUG------CCCC-------UAUCCUGCGCCCGUCACGGCCAAAUCCGUGACCGGGUUUGACAGCCCGAA--GAGAC-G-GCGCAAUG-CGCCGAGCGGU-GCAUGCGAAAGCAGCGA ..((------(...-------.....((((((.(((((((......)))))))(((((......)))))..--.....-)-)))))(((-((((....)))-))))((....)).))). ( -47.00) >pau.2265 2251612 100 + 6537648 CCUG------CCCC-------UAUCCUGCGCCCGUCACGGCCAAAUCCGUGACCGGGUUUGACAGCCCGAA--GAGAC-G-GCGCGAUG-CGCCGAGCGGU-GCAUGCGAAAGCAGCGA ..((------(...-------.....((((((.(((((((......)))))))(((((......)))))..--.....-)-)))))(((-((((....)))-))))((....)).))). ( -46.40) >ppk.369 5422293 101 + 6181863 CAUU-GCAGCCCCCACGACGCUACCGUGCUCCAGUCGCGGCCAACUCCGCGACCGGGUUUGGC-GACCCGAACGAAAA-G-GCGCAUCGGCGCC-----CCCGACCACGA--------- ....-..(((.........)))..((((.....(((((((......)))))))(((((((((.-...)))))).....-(-((((....)))))-----.)))..)))).--------- ( -36.20) >pel.378 791124 101 - 5888780 CAUAAGCAGGCCCUAAGGAAUAAAUUCCCUCC-GUCGCGGCCAUUUCCGCAACCGGGUUUGGC-GACCCGACCGAAAAAG-GCGCAACGGCGCC-----CC-GACCACGA--------- ........((......((((....)))).((.-(((((((......))))....(((((....-)))))))).))....(-((((....)))))-----..-..))....--------- ( -29.90) >psp.2360 1297201 94 + 5928787 GCUAUUCAGGUCUC-------UAGCCUUCAACGGUCGCUGCCCAAUCAGCGAUCAGGUUUGGUAGACCUGCUUUUGCU-G-GCGCAAUG-UGCC-----AC-CUCUGCGA--------- ((....((((((((-------((((((.....((((((((......))))))))))).)))).))))))).....))(-(-((((...)-))))-----).-........--------- ( -36.00) >pss.2449 1265803 95 + 6093698 GCUAUUCAGGUCUC-------UAGCCUUCAUCGGUCGCUGCCCAAUCAGCGAUCAGGUUUGGUAGACCUGAGUGACCU-GUGCGCAAUG-CGCC-----AC-CUUCUCAA--------- ..((((((((((((-------((((((.....((((((((......))))))))))).)))).)))))))))))...(-(.((((...)-))))-----).-........--------- ( -37.50) >consensus CCUA__CAGGCCCC_______UAGCCUGCACCCGUCGCGGCCAAAUCCGCGACCGGGUUUGGCAGACCCGA__GAGAA_G_GCGCAAUG_CGCC_____AC_GACCGCGA_________ .................................(((((((......)))))))((((((.....))))))...........(((......))).......................... (-16.60 = -16.30 + -0.30)

| Location | 1,348,596 – 1,348,699 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 56.01 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -14.64 |
| Energy contribution | -13.83 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.758 1348596 103 + 6264404 -GUCUC--UUCGGGCUGUCAAACCCGGUCACGGAUUUGGCCGUGACGGGCGCAGGAUA-------GGGG------CAGGCAAGGCGGGGAGCAAGGGCGGGAACGGGUGACGGCAGAGG -....(--(((..((((((..((((((((((((......)))))))..((.(......-------.).)------)..((...((.....))....)).....))))))))))).)))) ( -40.10) >pau.2265 2251649 103 - 6537648 -GUCUC--UUCGGGCUGUCAAACCCGGUCACGGAUUUGGCCGUGACGGGCGCAGGAUA-------GGGG------CAGGCAAGGCGGGGAGCAAGGGCGGGAACGGGUGACGGCAGAGG -....(--(((..((((((..((((((((((((......)))))))..((.(......-------.).)------)..((...((.....))....)).....))))))))))).)))) ( -40.10) >ppk.369 5422318 110 - 6181863 -UUUUCGUUCGGGUC-GCCAAACCCGGUCGCGGAGUUGGCCGCGACUGGAGCACGGUAGCGUCGUGGGGGCUGC-AAUGCAAGGUUUGGGG---GGAUGCGAAUGGA---CGCUGGGAG -..((((((((.(((-.((((((((((((((((......)))))))))..(((..(((((.(.....).)))))-..)))..)))))))..---.))).))))))))---......... ( -52.50) >pel.378 791148 111 + 5888780 UUUUUCGGUCGGGUC-GCCAAACCCGGUUGCGGAAAUGGCCGCGAC-GGAGGGAAUUUAUUCCUUAGGGCCUGCUUAUGCAAGCUGGGGAU---GGCUGGGAAAAGG---CAACGGACG (((..(((((..(((-.(((..(((.(((((((......)))))))-.)((((((....)))))).))((.(((....))).))))).)))---)))))..))).(.---...)..... ( -41.90) >psp.2360 1297224 92 - 5928787 -AGCAAAAGCAGGUCUACCAAACCUGAUCGCUGAUUGGGCAGCGACCGUUGAAGGCUA-------GAGACCUGAAUAGCCAAGC---------------GCAAGAGG---CCG-GGAUU -........(((((((.((...((..(((...)))..))((((....))))..))...-------.)))))))....(((...(---------------....).))---)..-..... ( -27.30) >pss.2449 1265827 92 - 6093698 -AGGUCACUCAGGUCUACCAAACCUGAUCGCUGAUUGGGCAGCGACCGAUGAAGGCUA-------GAGACCUGAAUAGCCAAGC---------------GCAAGAGG---CCG-GGAUU -.((((..((((((((.((.....((.((((((......)))))).)).....))...-------.)))))))).........(---------------....).))---)).-..... ( -30.20) >consensus _AUCUC__UCAGGUCUGCCAAACCCGGUCGCGGAUUUGGCCGCGACCGGAGCAGGAUA_______GGGGCCUG__UAGGCAAGC_GGGGAG___GG__GGGAACAGG___CGGCGGAGG .........((((.........))))(((((((......)))))))......................................................................... (-14.64 = -13.83 + -0.80)

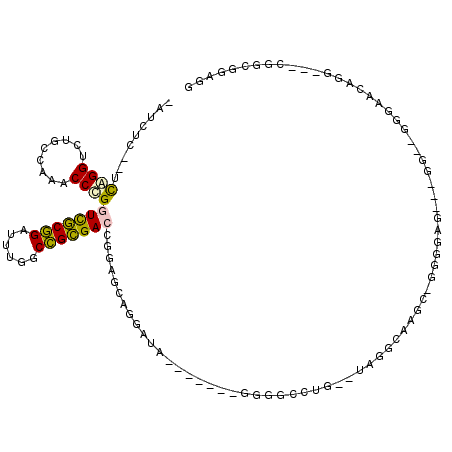
| Location | 1,348,596 – 1,348,699 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 56.01 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.758 1348596 103 - 6264404 CCUCUGCCGUCACCCGUUCCCGCCCUUGCUCCCCGCCUUGCCUG------CCCC-------UAUCCUGCGCCCGUCACGGCCAAAUCCGUGACCGGGUUUGACAGCCCGAA--GAGAC- .((((((.((((.........((....((.....))...))...------....-------........(((((((((((......)))))).))))).)))).))....)--)))..- ( -27.50) >pau.2265 2251649 103 + 6537648 CCUCUGCCGUCACCCGUUCCCGCCCUUGCUCCCCGCCUUGCCUG------CCCC-------UAUCCUGCGCCCGUCACGGCCAAAUCCGUGACCGGGUUUGACAGCCCGAA--GAGAC- .((((((.((((.........((....((.....))...))...------....-------........(((((((((((......)))))).))))).)))).))....)--)))..- ( -27.50) >ppk.369 5422318 110 + 6181863 CUCCCAGCG---UCCAUUCGCAUCC---CCCCAAACCUUGCAUU-GCAGCCCCCACGACGCUACCGUGCUCCAGUCGCGGCCAACUCCGCGACCGGGUUUGGC-GACCCGAACGAAAA- ......(((---......)))....---.(((((((((.((((.-(.(((.........))).).))))....(((((((......))))))).)))))))).-).............- ( -32.90) >pel.378 791148 111 - 5888780 CGUCCGUUG---CCUUUUCCCAGCC---AUCCCCAGCUUGCAUAAGCAGGCCCUAAGGAAUAAAUUCCCUCC-GUCGCGGCCAUUUCCGCAACCGGGUUUGGC-GACCCGACCGAAAAA .(((.((((---((....(((....---.(((...((((((....)))))).....))).............-((.((((......)))).)).)))...)))-)))..)))....... ( -34.40) >psp.2360 1297224 92 + 5928787 AAUCC-CGG---CCUCUUGC---------------GCUUGGCUAUUCAGGUCUC-------UAGCCUUCAACGGUCGCUGCCCAAUCAGCGAUCAGGUUUGGUAGACCUGCUUUUGCU- .....-.((---((......---------------....))))...((((((((-------((((((.....((((((((......))))))))))).)))).)))))))........- ( -31.80) >pss.2449 1265827 92 + 6093698 AAUCC-CGG---CCUCUUGC---------------GCUUGGCUAUUCAGGUCUC-------UAGCCUUCAUCGGUCGCUGCCCAAUCAGCGAUCAGGUUUGGUAGACCUGAGUGACCU- .....-.((---((....).---------------))).((.((((((((((((-------((((((.....((((((((......))))))))))).)))).))))))))))).)).- ( -36.60) >consensus CAUCCGCCG___CCCCUUCCC__CC___CUCCCC_CCUUGCCUA__CAGGCCCC_______UAGCCUGCACCCGUCGCGGCCAAAUCCGCGACCGGGUUUGGCAGACCCGA__GAGAA_ .........................................................................(((((((......)))))))((((((.....))))))......... (-14.28 = -14.12 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:18:24 2007