
| Sequence ID | pao.917 |
|---|---|
| Location | 1,644,820 – 1,644,981 |
| Length | 161 |
| Max. P | 0.999991 |

| Location | 1,644,820 – 1,644,931 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -26.36 |
| Energy contribution | -25.81 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644820 111 + 6264404 ---UCAAGGCGUUAUUGGGGGAAGCCAAAAAUUCGCUGGCGAUUCUCAUGGGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCG ---.............((((((.((((.........))))..((((....)))).....------........((((((((((((((((.......)))))))))))))))).)))))). ( -37.20) >pau.2111 2537654 111 - 6537648 ---UCAAGGCGUUAUUGGGGGAAGCCAAAAACUCGCUGGCGAUUCUCAUGGGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCG ---.............((((((.((((.........))))..((((....)))).....------........((((((((((((((((.......)))))))))))))))).)))))). ( -37.20) >pel.3041 5594687 114 + 5888780 CUUUCAA----UUACUUAAAAACGUCACGCAUUCAUUUG-GAUGCUC-UAGAAAGAAAAAACCAUCCAAAAGUUGCGCAACUUUUUGCGCAGUGGUGGCAAAAAGUUGCGCAGUUGGCUG (((((..----..((........))...((((((....)-)))))..-..))))).......((.((((...((((((((((((((((.((....)))))))))))))))))))))).)) ( -37.50) >consensus ___UCAAGGCGUUAUUGGGGGAAGCCAAAAAUUCGCUGGCGAUUCUCAUGGGAAGAACA______CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCG ................(((((...((......(((....)))........)).....................(((((((((((((((.........))))))))))))))))))))... (-26.36 = -25.81 + -0.55)

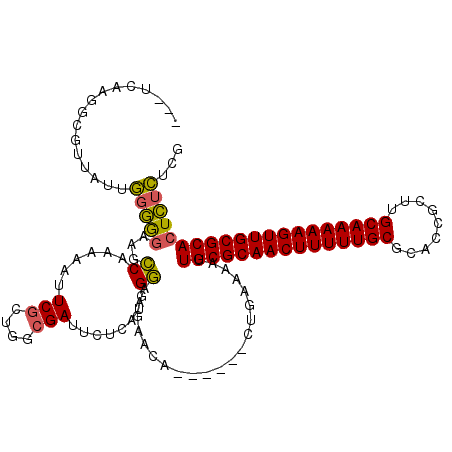
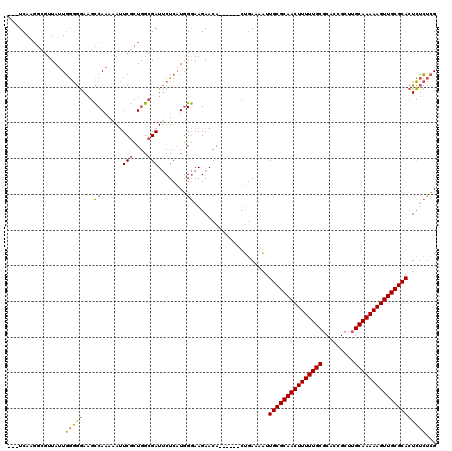
| Location | 1,644,820 – 1,644,931 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -26.81 |
| Energy contribution | -26.27 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 5.61 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644820 111 - 6264404 CGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCCCAUGAGAAUCGCCAGCGAAUUUUUGGCUUCCCCCAAUAACGCCUUGA--- ...((((((((((((((((((((.........)))))))))))))).)))))).(------(((((((.....))))).)))..(((.....((((......))))...))).....--- ( -36.70) >pau.2111 2537654 111 + 6537648 CGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCCCAUGAGAAUCGCCAGCGAGUUUUUGGCUUCCCCCAAUAACGCCUUGA--- ...((((((((((((((((((((.........)))))))))))))).)))))).(------(((((((.....))))).)))(((.(.(((.((((......)))).))).).))).--- ( -37.90) >pel.3041 5594687 114 - 5888780 CAGCCAACUGCGCAACUUUUUGCCACCACUGCGCAAAAAGUUGCGCAACUUUUGGAUGGUUUUUUCUUUCUA-GAGCAUC-CAAAUGAAUGCGUGACGUUUUUAAGUAA----UUGAAAG ..((....(((((((((((((((((....)).))))))))))))))).(.((((((((.(((.........)-)).))))-)))).)...)).......((((((....----)))))). ( -38.90) >consensus CGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG______UGUUCUUCCCAUGAGAAUCGCCAGCGAAUUUUUGGCUUCCCCCAAUAACGCCUUGA___ ((((....(((((((((((((((.........)))))))))))))))......................(((...((((((....)).)))).)))................)))).... (-26.81 = -26.27 + -0.54)

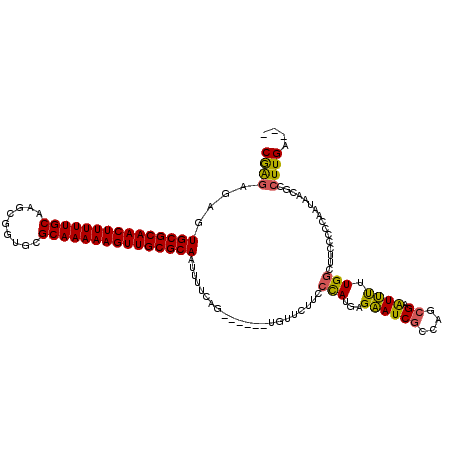
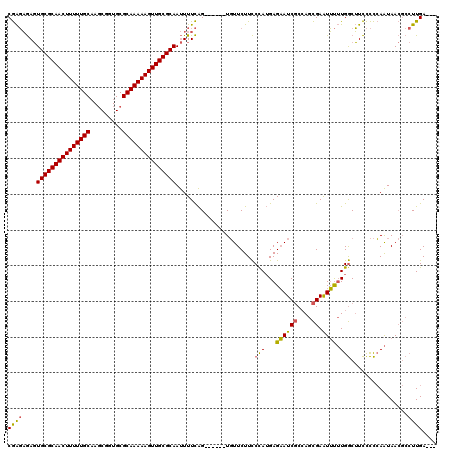
| Location | 1,644,857 – 1,644,971 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.97 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.70 |
| SVM decision value | 5.17 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644857 114 + 6264404 GAUUCUCAUGGGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAA ........((((.(((((.------((((....((((((((((((((((.......)))))))))))))))).....)))).((.....(((........)))..))))))).))))... ( -41.90) >pau.2111 2537691 114 - 6537648 GAUUCUCAUGGGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAA ........((((.(((((.------((((....((((((((((((((((.......)))))))))))))))).....)))).((.....(((........)))..))))))).))))... ( -41.90) >pel.3041 5594722 119 + 5888780 GAUGCUC-UAGAAAGAAAAAACCAUCCAAAAGUUGCGCAACUUUUUGCGCAGUGGUGGCAAAAAGUUGCGCAGUUGGCUGUAGGCCUUGCUGCGCAUGCUCUACAUCAAUGUAUUCACAC ((((...-((((.........(((.(((....((((((((....))))))))))))))......((((((((((.(((.....)))..)))))))).))))))))))............. ( -39.80) >consensus GAUUCUCAUGGGAAGAACA______CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAA ..........((((((((.......((((....(((((((((((((((.........))))))))))))))).....)))).(((.....)))..............))))).))).... (-28.96 = -29.97 + 1.01)

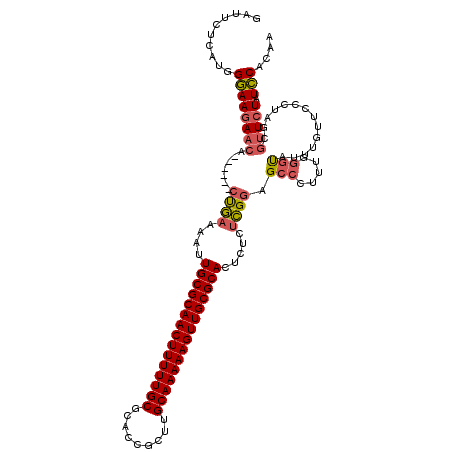
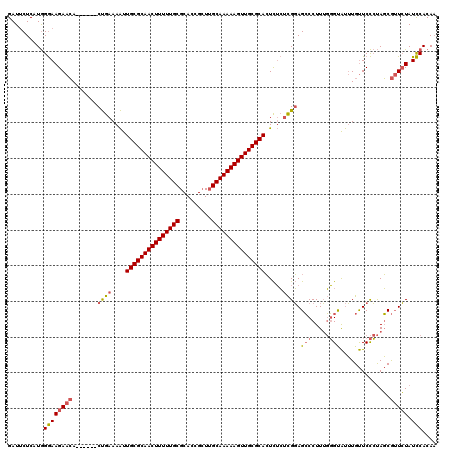
| Location | 1,644,857 – 1,644,971 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -45.90 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644857 114 - 6264404 UUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCCCAUGAGAAUC (..(((..((((((((.(((........)))............((((((((((((((((((((.........)))))))))))))).))))))))------))))))..)))..)..... ( -47.90) >pau.2111 2537691 114 + 6537648 UUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCCCAUGAGAAUC (..(((..((((((((.(((........)))............((((((((((((((((((((.........)))))))))))))).))))))))------))))))..)))..)..... ( -47.90) >pel.3041 5594722 119 - 5888780 GUGUGAAUACAUUGAUGUAGAGCAUGCGCAGCAAGGCCUACAGCCAACUGCGCAACUUUUUGCCACCACUGCGCAAAAAGUUGCGCAACUUUUGGAUGGUUUUUUCUUUCUA-GAGCAUC (((((..((((....))))...)))))((.....(((.....)))...(((((((((((((((((....)).)))))))))))))))...((((((.((......)).))))-))))... ( -41.90) >consensus UUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG______UGUUCUUCCCAUGAGAAUC .((.(((.((((((...(((........)))..........((((...(((((((((((((((.........)))))))))))))))..))))........)))))))))))........ (-29.70 = -29.93 + 0.23)

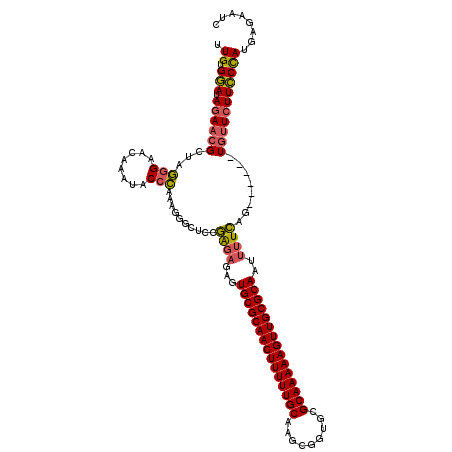
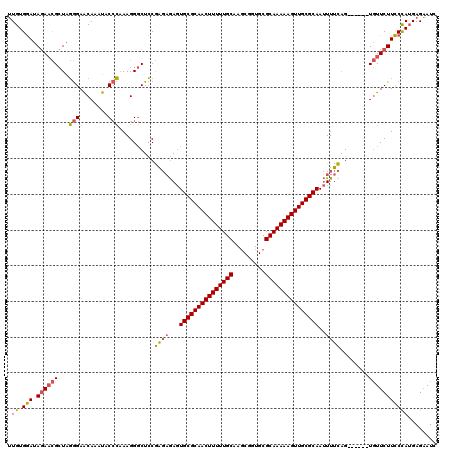
| Location | 1,644,867 – 1,644,981 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -28.89 |
| Energy contribution | -29.90 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.72 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644867 114 + 6264404 GGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAACGUUAUCAAC ((((((((.------((((....((((((((((((((((.......)))))))))))))))).....)))).((.....(((........)))..))))))).))).............. ( -40.90) >pau.2111 2537701 114 - 6537648 GGAAGAACA------CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAACGUUAUCAAC ((((((((.------((((....((((((((((((((((.......)))))))))))))))).....)))).((.....(((........)))..))))))).))).............. ( -40.90) >pel.3041 5594731 120 + 5888780 GAAAGAAAAAACCAUCCAAAAGUUGCGCAACUUUUUGCGCAGUGGUGGCAAAAAGUUGCGCAGUUGGCUGUAGGCCUUGCUGCGCAUGCUCUACAUCAAUGUAUUCACACACUUAUCCAC ....(((................(((((((((((((((.((....)))))))))))))))))(((((.(((((((..(((...))).)).))))))))))...))).............. ( -37.90) >consensus GGAAGAACA______CUGAAAAUUGCGCAACUUUUUGCGCACCGCUUGCAAAAAGUUGCGCACUCUCUCGGAGCCCUUUGGGUAUUUGUUCCCUAGCGUUCUAUCCACAACGUUAUCAAC ((((((((.......((((....(((((((((((((((.........))))))))))))))).....)))).(((.....)))..............))))).))).............. (-28.89 = -29.90 + 1.01)

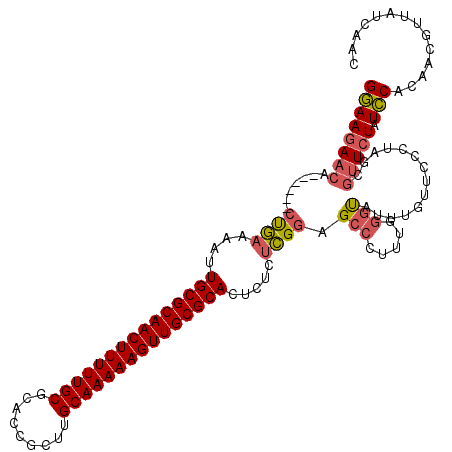
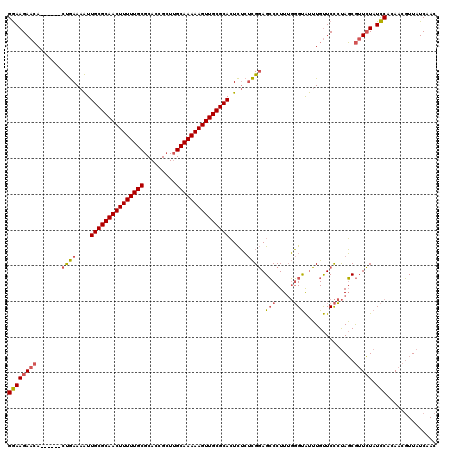
| Location | 1,644,867 – 1,644,981 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.917 1644867 114 - 6264404 GUUGAUAACGUUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCC ..............(((.((((((((.(((........)))............((((((((((((((((((((.........)))))))))))))).))))))))------))))))))) ( -43.50) >pau.2111 2537701 114 + 6537648 GUUGAUAACGUUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG------UGUUCUUCC ..............(((.((((((((.(((........)))............((((((((((((((((((((.........)))))))))))))).))))))))------))))))))) ( -43.50) >pel.3041 5594731 120 - 5888780 GUGGAUAAGUGUGUGAAUACAUUGAUGUAGAGCAUGCGCAGCAAGGCCUACAGCCAACUGCGCAACUUUUUGCCACCACUGCGCAAAAAGUUGCGCAACUUUUGGAUGGUUUUUUCUUUC ........(((((((..((((....))))...)))))))...((((((.....((((.(((((((((((((((((....)).)))))))))))))))....))))..))))))....... ( -42.30) >consensus GUUGAUAACGUUGUGGAUAGAACGCUAGGGAACAAAUACCCAAAGGGCUCCGAGAGAGUGCGCAACUUUUUGCAAGCGGUGCGCAAAAAGUUGCGCAAUUUUCAG______UGUUCUUCC ..............(((.((((((...(((........)))..........((((...(((((((((((((((.........)))))))))))))))..))))........))))))))) (-28.55 = -29.00 + 0.45)

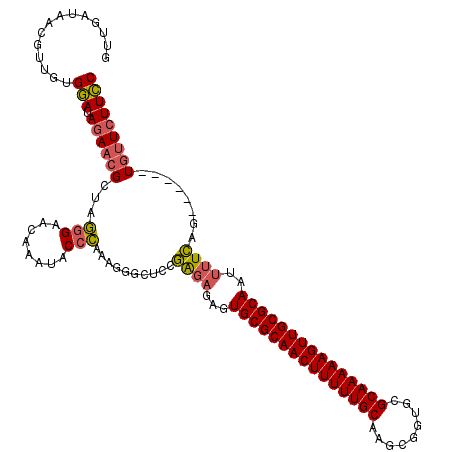
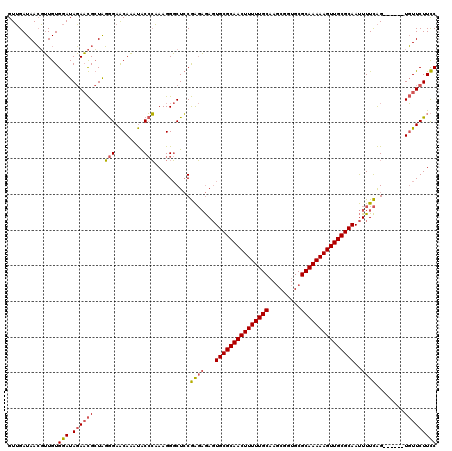
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:18:09 2007