| Sequence ID | pao.345 |
|---|---|
| Location | 605,944 – 606,054 |
| Length | 110 |
| Max. P | 0.994317 |

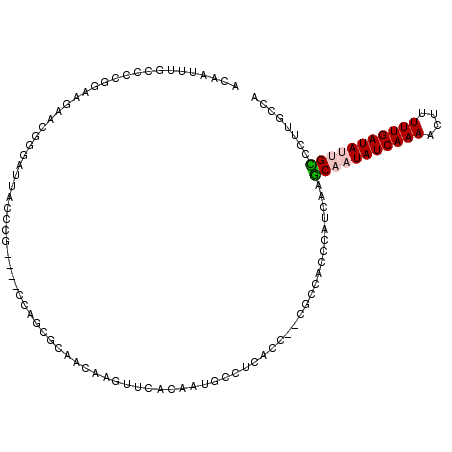
| Location | 605,944 – 606,054 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.22 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -15.24 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.345 605944 110 + 6264404 CGGCAAUGCCUGUAUCAAAAAUUUUUGAUAUUGA-UGACGGGUGGCC--GUCGCGCAAUUGCAAUCCGGCUGCGCCAGGCUUCUGGCAAAACCGGUCGGCAGCGACAUACUGU .(((...(((((((((((............))))-).)))))).)))--(((((((....))...(((((((.(((((....))))).....)))))))..)))))....... ( -43.00) >pau.348 615920 110 + 6537648 CGGCAAUGCCUGUAUCAAAAAUUUUUGAUAUUGA-UGACGGGUGGCC--GUCGCGCAAUUGCAAUCCGGCUGCGCCAGGCUUCUGGCAAAACCGGUCGGCAGCGACAUACUGU .(((...(((((((((((............))))-).)))))).)))--(((((((....))...(((((((.(((((....))))).....)))))))..)))))....... ( -43.00) >pel.2885 551807 106 - 5888780 UAGCAAGACCAAUAUCAAAAAGUUUUGAUAUUGGUCGAUGGGCGGAG--GAUGGGUUUGCCUGUGCGGGCCUCUUUG-----CGGGCAGUUCCGCUGUUUUCACCCAAAUUGC ..(((((((((((((((((....)))))))))))))..((((.((((--(.(((..(((((((((.((....)).))-----)))))))..)))...))))).))))..)))) ( -46.20) >pst.243 425770 109 + 6397126 UCGCAAGGGCAAUAUCAAAAAGUUUUGAUAUUGCUUGAUGGGUGGCAUCGGUCAGGCGUUGUGAACUUGUCGCGCUGA----UGGGUAAUCCAGUUCUUACAGACCAAAUAAU ......(((((((((((((....)))))))))))....(((((...(((.((((((((..(....)....))).))))----).))).)))))...........))....... ( -33.80) >psp.2808 454414 109 - 5928787 CUGCAAGGGCAAUAUCAAAAAGUUUUGAUAUUGCUGGAUGGGUGGCACCGGUCAGGCAUUCUGAACUUGUUGCGCUGA----CGGGUAAUCCAGUCUUUACAGGGCAAAUAAU .(((...((((((((((((....))))))))))))((((((((...(((.((((((((............))).))))----).))).))))).))).......)))...... ( -38.60) >pss.2907 412006 109 - 6093698 UCGCAAGGGCAAUAUCAAAAAGUUUUGAUAUUGCGGGAUGGAUGGCGUCGGUCAGGCGUUGUGAACUUGUCGUGCAGG----CGGGUGAUCCAGUCUUUGCGUGGCAAAUAAU .((((((((((((((((((....))))))))))).((((..(..(((((.....)))))..)..(((((((.....))----))))).))))...)))))))........... ( -42.00) >consensus CCGCAAGGCCAAUAUCAAAAAGUUUUGAUAUUGCUUGAUGGGUGGCA__GGUCAGGCAUUGUGAACUGGCCGCGCUGG____CGGGCAAUCCAGUUCUUACGCGACAAAUAAU ..((....(((((((((((....))))))))))).....(.((((((....................)))))).)..........)).......................... (-15.24 = -14.47 + -0.78)

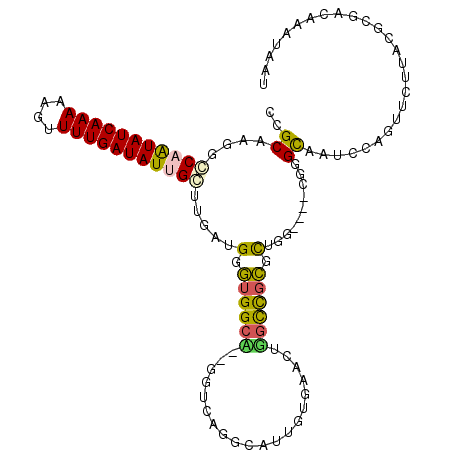
| Location | 605,944 – 606,054 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 63.22 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.58 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.345 605944 110 - 6264404 ACAGUAUGUCGCUGCCGACCGGUUUUGCCAGAAGCCUGGCGCAGCCGGAUUGCAAUUGCGCGAC--GGCCACCCGUCA-UCAAUAUCAAAAAUUUUUGAUACAGGCAUUGCCG .((((.....))))....((((((.((((((....)))))).))))))...(((((.((..(((--((....))))).-....(((((((....)))))))...))))))).. ( -40.70) >pau.348 615920 110 - 6537648 ACAGUAUGUCGCUGCCGACCGGUUUUGCCAGAAGCCUGGCGCAGCCGGAUUGCAAUUGCGCGAC--GGCCACCCGUCA-UCAAUAUCAAAAAUUUUUGAUACAGGCAUUGCCG .((((.....))))....((((((.((((((....)))))).))))))...(((((.((..(((--((....))))).-....(((((((....)))))))...))))))).. ( -40.70) >pel.2885 551807 106 + 5888780 GCAAUUUGGGUGAAAACAGCGGAACUGCCCG-----CAAAGAGGCCCGCACAGGCAAACCCAUC--CUCCGCCCAUCGACCAAUAUCAAAACUUUUUGAUAUUGGUCUUGCUA ((((..((((((......((((......)))-----)......(((......))).........--...))))))..(((((((((((((....))))))))))))))))).. ( -40.90) >pst.243 425770 109 - 6397126 AUUAUUUGGUCUGUAAGAACUGGAUUACCCA----UCAGCGCGACAAGUUCACAACGCCUGACCGAUGCCACCCAUCAAGCAAUAUCAAAACUUUUUGAUAUUGCCCUUGCGA ...........((((((...(((.....)))----((((.(((............)))))))..((((.....))))..(((((((((((....))))))))))).)))))). ( -27.00) >psp.2808 454414 109 + 5928787 AUUAUUUGCCCUGUAAAGACUGGAUUACCCG----UCAGCGCAACAAGUUCAGAAUGCCUGACCGGUGCCACCCAUCCAGCAAUAUCAAAACUUUUUGAUAUUGCCCUUGCAG ..........((((((....(((((((((.(----((((.(((............)))))))).))))......)))))(((((((((((....)))))))))))..)))))) ( -34.00) >pss.2907 412006 109 + 6093698 AUUAUUUGCCACGCAAAGACUGGAUCACCCG----CCUGCACGACAAGUUCACAACGCCUGACCGACGCCAUCCAUCCCGCAAUAUCAAAACUUUUUGAUAUUGCCCUUGCGA ...........(((((....(((.(((..((----..((.((.....)).))...))..))))))..............(((((((((((....)))))))))))..))))). ( -23.80) >consensus ACAAUUUGCCCCGGAAGAACGGGAUUACCCG____CCAGCGCAACAAGUUCACAAUGCCUCACC__CGCCACCCAUCAAGCAAUAUCAAAACUUUUUGAUAUUGCCCUUGCCA ...............................................................................(((((((((((....)))))))))))........ ( -9.47 = -9.58 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:18:00 2007