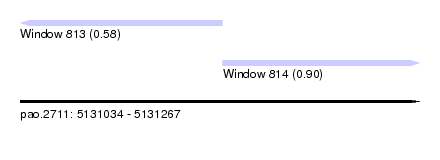
| Sequence ID | pao.2711 |
|---|---|
| Location | 5,131,034 – 5,131,267 |
| Length | 233 |
| Max. P | 0.895348 |

| Location | 5,131,034 – 5,131,152 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.50 |
| Mean single sequence MFE | -45.97 |
| Consensus MFE | -19.03 |
| Energy contribution | -21.43 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2711 5131034 118 - 6264404 UCGAGCGGGAGUCGAACCCGCGACAGACCAUUUCGUGGAUGGUUGCUCUACCAGGCUGAGCUAUCGAACCGCGAAGCGGCAUUGCCGUCAUUCGCGAAUGACCGUCGUGGUUGCGG--GC ((((.(....))))).((((((((...((.((((((((.((((.((((.........)))).))))..)))))))).))(((.((.((((((....)))))).)).))))))))))--). ( -49.10) >pau.2848 5403501 118 - 6537648 UCGAGCGGGAGUCGAACCCGCGACAGACCAUUUCGUGGAUGGUUGCUCUACCAGGCUGAGCUAUCGAACCGCGAAGCGGCAUUGCCGUCAUUCGCGAAUGACCGUCGUUGUUGCGG--GC ((((.(....))))).((((((((((..(((...)))(((((((((((.........))))........(((((((((((...)))))..))))))...))))))).)))))))))--). ( -48.90) >pel.297 625119 74 - 5888780 CGGGACAGGGCCCGAC---------GAGCGCCUCGC--AAGCCUGUUCAA--AGGCU-------UGCGCCGCAAAG------------------------ACCG--GUCGUUGCGGCGGC ((((......))))..---------....(((..((--((((((......--)))))-------)))(((((((.(------------------------(...--.)).)))))))))) ( -39.90) >consensus UCGAGCGGGAGUCGAACCCGCGACAGACCAUUUCGUGGAUGGUUGCUCUACCAGGCUGAGCUAUCGAACCGCGAAGCGGCAUUGCCGUCAUUCGCGAAUGACCGUCGUCGUUGCGG__GC ..((((((..((((......))))...(((((.....)))))))))))....................((((((.(((((......((((((....)))))).)))))..)))))).... (-19.03 = -21.43 + 2.40)

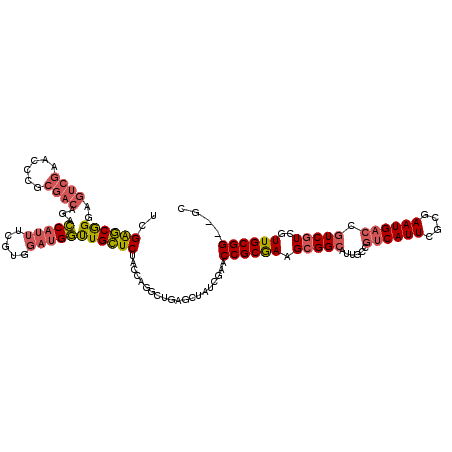
| Location | 5,131,152 – 5,131,267 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -48.43 |
| Consensus MFE | -35.14 |
| Energy contribution | -34.03 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2711 5131152 115 + 6264404 ACGUUGCC--UUAACGGCAAUCACUCAAUGGCAACACCCGCCGGUGUGGCCGAGGCGAACUGCGCAAGC--GGCCAGGAAGGCCGGACAACGCAAGGGA-GCCGAAGCCGCCGCCGGCCC ..((((((--.....((......))....))))))....(((((((((((...(((...(((((....(--((((.....))))).....))).))...-)))...)))).))))))).. ( -50.60) >pau.2848 5403619 115 + 6537648 ACGUUGCC--UUAACGGCAAUCACUCAAUGGCAACACCCGCCGGUGUGGCCGAGGCGAACUGCGCAAGC--GGCCAGGAAGGCCGGACAACGCAAGGGA-GCCGAAGCCGCCGCCGGCCC ..((((((--.....((......))....))))))....(((((((((((...(((...(((((....(--((((.....))))).....))).))...-)))...)))).))))))).. ( -50.60) >pel.297 625193 114 + 5888780 ACACUACCCUUCCACGGUAGCUA--CAAACGCAACACCCGCAGGUGGGGCACAGGCAA----CGCAACCAGGGCCAGGACGGUCCGCAUUUGCGAAGGACGCCUGGGCCACCGCUGGCCC .......(((((...(((.((..--.....((..((((....))))..))...(....----))).))).(((((.....)))))((....)))))))......((((((....)))))) ( -44.10) >consensus ACGUUGCC__UUAACGGCAAUCACUCAAUGGCAACACCCGCCGGUGUGGCCGAGGCGAACUGCGCAAGC__GGCCAGGAAGGCCGGACAACGCAAGGGA_GCCGAAGCCGCCGCCGGCCC ...(((((.......)))))...................(((((((((((...(((...............((((.....))))......(....)....)))...)))).))))))).. (-35.14 = -34.03 + -1.10)

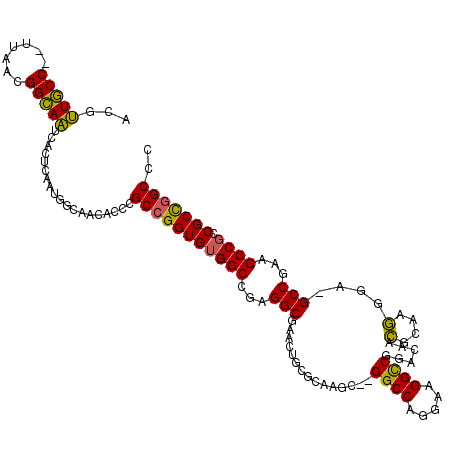

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:58 2007