
| Sequence ID | pao.2750 |
|---|---|
| Location | 5,203,376 – 5,203,667 |
| Length | 291 |
| Max. P | 0.999993 |

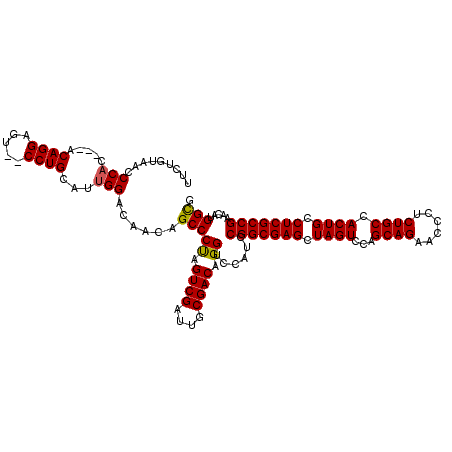
| Location | 5,203,376 – 5,203,491 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -36.11 |
| Energy contribution | -36.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203376 115 + 6264404 UUCUGUAACCCAC---ACAGGAGU--CCUGCAUUGGACAACAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCG ((((((.......---))))))((--((......))))......(((.((((....))))))).((((.(((((((.((((...(((((....))))).)))).)))))))...)))).. ( -42.90) >pau.2886 5475355 115 + 6537648 UUCUGUAACCCAC---ACAGGAGU--CCUGCAUUGGACAACAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCG ((((((.......---))))))((--((......))))......(((.((((....))))))).((((.(((((((.((((...(((((....))))).)))).)))))))...)))).. ( -42.90) >pel.1792 3328656 119 + 5888780 CUCAAGCAUCCCUUCAACAGGAGUCCCCUGCAUUGGACAGUAGCCCCAGUCGAUUGCGACAGGCCCACACGGCGAGCUAGUCCGGCAGCGUUC-CUGCCACUGUCUCGCCGCACCCGGUA .................((((.....)))).(((((...((.(((...((((....)))).)))..)).(((((((.((((..(((((.....-))))))))).)))))))...))))). ( -40.30) >consensus UUCUGUAACCCAC___ACAGGAGU__CCUGCAUUGGACAACAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCG .........(((.....((((.....))))...)))......(((((.((((....)))).))......(((((((.((((...((((......)))).)))).))))))).....))). (-36.11 = -36.00 + -0.11)

| Location | 5,203,376 – 5,203,491 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -46.65 |
| Energy contribution | -45.67 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203376 115 - 6264404 CGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUGUUGUCCAAUGCAGG--ACUCCUGU---GUGGGUUACAGAA ..(((..(((((((((..((((((((......)))))..)))..)))))))))))).(((((((....)))).))).((((..((((.((((((--...)))))---)))))..)))).. ( -49.10) >pau.2886 5475355 115 - 6537648 CGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUGUUGUCCAAUGCAGG--ACUCCUGU---GUGGGUUACAGAA ..(((..(((((((((..((((((((......)))))..)))..)))))))))))).(((((((....)))).))).((((..((((.((((((--...)))))---)))))..)))).. ( -49.10) >pel.1792 3328656 119 - 5888780 UACCGGGUGCGGCGAGACAGUGGCAG-GAACGCUGCCGGACUAGCUCGCCGUGUGGGCCUGUCGCAAUCGACUGGGGCUACUGUCCAAUGCAGGGGACUCCUGUUGAAGGGAUGCUUGAG ...(((((((((((((..((((((((-.....)))))..)))..))))))))((((.(((((((....)))).))).)))).((((...(((((.....))))).....))))))))).. ( -53.60) >consensus CGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUGUUGUCCAAUGCAGG__ACUCCUGU___GUGGGUUACAGAA ..(((..(((((((((..((((((((......)))))..)))..)))))))))))).(((((((....)))).))).((((..((((..(((((.....)))))....))))..)))).. (-46.65 = -45.67 + -0.99)

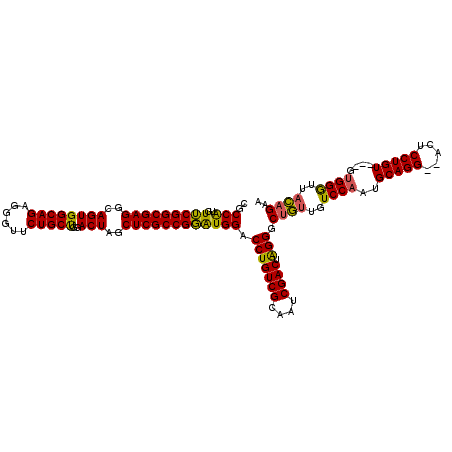

| Location | 5,203,411 – 5,203,531 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -50.33 |
| Consensus MFE | -44.35 |
| Energy contribution | -43.13 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203411 120 + 6264404 CAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCC ..(((((.((((....))))..((((..((((((((.((((...(((((....))))).)))).))))))).....)..)))))).)))(((.((((((((....)))))).)).))).. ( -52.70) >pau.2886 5475390 120 + 6537648 CAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCC ..(((((.((((....))))..((((..((((((((.((((...(((((....))))).)))).))))))).....)..)))))).)))(((.((((((((....)))))).)).))).. ( -52.70) >pel.1792 3328696 118 + 5888780 UAGCCCCAGUCGAUUGCGACAGGCCCACACGGCGAGCUAGUCCGGCAGCGUUC-CUGCCACUGUCUCGCCGCACCCGGUAGACGGUGGUUUCCUCCAGCCCUCCAUGGCUGA-GCACCCC ..(((...((((....)))).)))((((.(((((((.((((..(((((.....-))))))))).)))))))....((.....)))))).......(((((......))))).-....... ( -45.60) >consensus CAGCCCUAGUCGAUUGCGACAGGUCCAUCCGGCGAGCUAGUCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCC ..(((((.((((....))))..(((....(((((((.((((...((((......)))).)))).))))))).....)))....)).)))......((((((....))))))......... (-44.35 = -43.13 + -1.21)

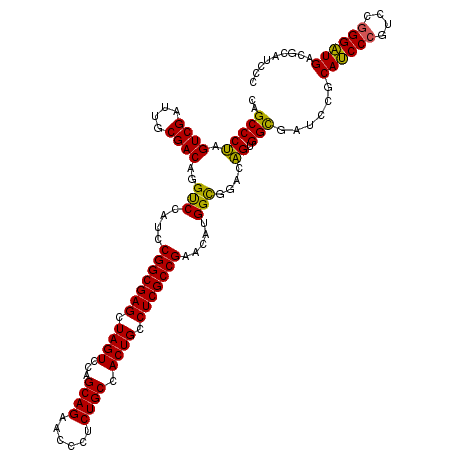
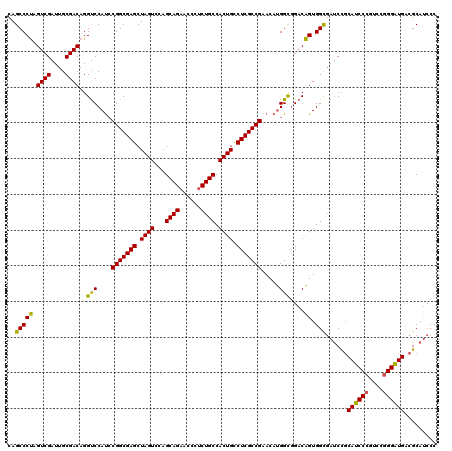
| Location | 5,203,411 – 5,203,531 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -57.27 |
| Consensus MFE | -48.65 |
| Energy contribution | -48.33 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203411 120 - 6264404 GGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUG (((((.((.((((((....)))))))).)))(((((((((((((......)))).))))))))).((((((((....)))))..))).))....((.(((((((....)))).))).)). ( -58.50) >pau.2886 5475390 120 - 6537648 GGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUG (((((.((.((((((....)))))))).)))(((((((((((((......)))).))))))))).((((((((....)))))..))).))....((.(((((((....)))).))).)). ( -58.50) >pel.1792 3328696 118 - 5888780 GGGGUGC-UCAGCCAUGGAGGGCUGGAGGAAACCACCGUCUACCGGGUGCGGCGAGACAGUGGCAG-GAACGCUGCCGGACUAGCUCGCCGUGUGGGCCUGUCGCAAUCGACUGGGGCUA ((..(.(-((((((......)))).))).)..)).(((.....)))(..(((((((..((((((((-.....)))))..)))..)))))))..).(((((((((....))))..))))). ( -54.80) >consensus GGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGACUAGCUCGCCGGAUGGACCUGUCGCAAUCGACUAGGGCUG (((((.((.((((((....)))))))).))).))...((((.(((..(((((((((..((((((((......)))))..)))..))))))))))))....((((....))))..)))).. (-48.65 = -48.33 + -0.32)

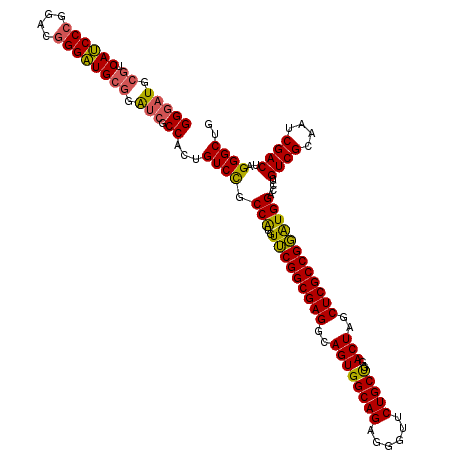

| Location | 5,203,451 – 5,203,571 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -31.41 |
| Energy contribution | -31.20 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 5.72 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203451 120 + 6264404 UCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAACCGCGCA ....(((((....)))))(((......(((......)))(((.(((((.(((.((((((((....)))))).)).))).))))).)))....)))...........((((.....)))). ( -43.90) >pau.2886 5475430 120 + 6537648 UCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAUCCGCGCA ....(((((....)))))(((......(((......)))(((.(((((.(((.((((((((....)))))).)).))).))))).)))....)))...........((((.....)))). ( -43.90) >pel.1792 3328736 108 + 5888780 UCCGGCAGCGUUC-CUGCCACUGUCUCGCCGCACCCGGUAGACGGUGGUUUCCUCCAGCCCUCCAUGGCUGA-GCACCCCUCUUUUCCAGCAG----CCUUGCCACCCG------GCAUG ...(((.((....-..((((((((((.((((....))))))))))))))......(((((......))))).-................)).)----)).((((....)------))).. ( -41.10) >consensus UCCAGCAGAACCCUCUGCCACUGCCUCGCCGAACAUGGCGGACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCA_CCGCGCA ....((..........(((((((.((.((((....)))))).)))))))......((((((....))))))..))...............................((((.....)))). (-31.41 = -31.20 + -0.21)

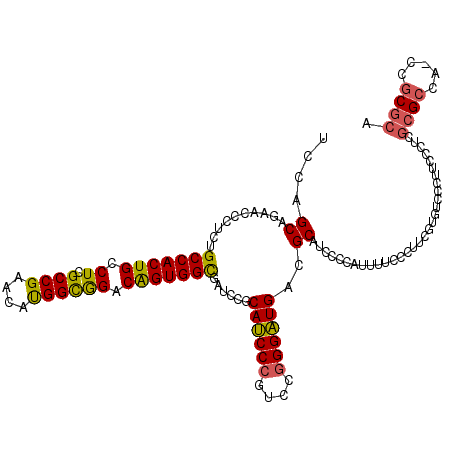
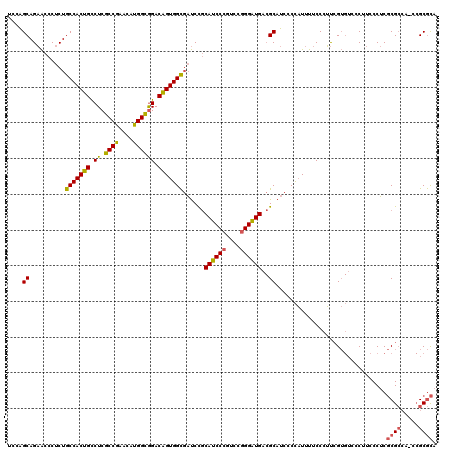
| Location | 5,203,451 – 5,203,571 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -35.33 |
| Energy contribution | -37.90 |
| Covariance contribution | 2.57 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203451 120 - 6264404 UGCGCGGUUGGCGCGAGGGAAGGGACACGAAGGGAAAAUGGGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGA .((((.....)))).......(((((................(((.((.((((((....)))))))).)))(((((((((((((......)))).))))))))).....)))))...... ( -54.30) >pau.2886 5475430 120 - 6537648 UGCGCGGAUGGCGCGAGGGAAGGGACACGAAGGGAAAAUGGGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGA .((((.....)))).......(((((................(((.((.((((((....)))))))).)))(((((((((((((......)))).))))))))).....)))))...... ( -54.30) >pel.1792 3328736 108 - 5888780 CAUGC------CGGGUGGCAAGG----CUGCUGGAAAAGAGGGGUGC-UCAGCCAUGGAGGGCUGGAGGAAACCACCGUCUACCGGGUGCGGCGAGACAGUGGCAG-GAACGCUGCCGGA ....(------((.(..((....----(((((........(((((.(-((((((......)))).)))...))).))((((.(((....)))..))))...)))))-....))..)))). ( -40.80) >consensus UGCGCGG_UGGCGCGAGGGAAGGGACACGAAGGGAAAAUGGGGAUGCGUCAUCCCGGACGGGAUGCGGAUCGCCACUGUCCGCCAUGUUCGGCGAGGCAGUGGCAGAGGGUUCUGCUGGA (((((.....))))).((...(((((.(..............(((.((.((((((....)))))))).)))(((((((((((((......)))).)))))))))...).))))).))... (-35.33 = -37.90 + 2.57)


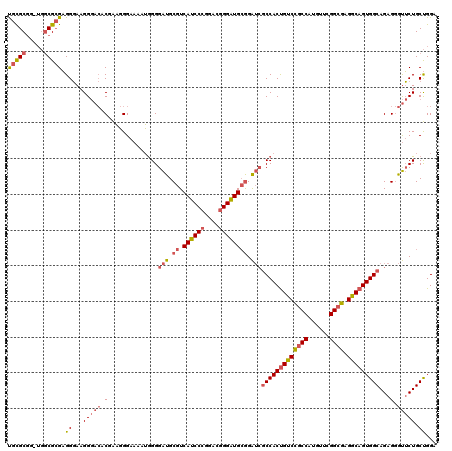
| Location | 5,203,491 – 5,203,611 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.28 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203491 120 + 6264404 GACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAACCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCCUUAUC ((((((((.(((.((((((((....)))))).)).))).)))))......(((((...((((((.(((((.....)))).).)))))).)))))................)))....... ( -43.10) >pau.2886 5475470 120 + 6537648 GACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAUCCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCUUCCUUUAUC ((((((((.(((.((((((((....)))))).)).))).))))).......((((...((((((.(((((.....)))).).)))))).)))))))........................ ( -42.40) >pel.1792 3328775 101 + 5888780 GACGGUGGUUUCCUCCAGCCCUCCAUGGCUGA-GCACCCCUCUUUUCCAGCAG----CCUUGCCACCCG------GCAUGGCG------CGUGGUCCUGCGCAUUCC-UCG-CUUUCAAU ((.((.(((...(((.((((......))))))-).)))))))......(((.(----((.((((....)------))).)))(------(((......)))).....-..)-))...... ( -34.40) >consensus GACAGUGGCGAUCCGCAUCCCGUCCGGGAUGACGCAUCCCCAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCA_CCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCUUUAUC ((.(((((.(((.((((((((....)))))).)).))).))))).))............................(((((((((....)))).....))))).................. (-24.38 = -25.03 + 0.65)


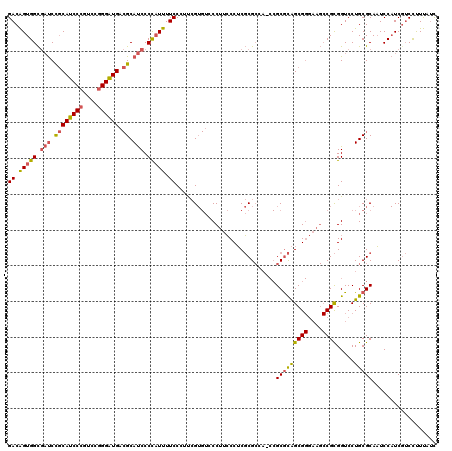
| Location | 5,203,531 – 5,203,651 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -47.74 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203531 120 + 6264404 CAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAACCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCCUUAUCCGCGCGGACCGUCGGCCCGCGCCAGCUCCGGCCGUCGGCC ...................(((((.(((((.....)))).).)))))((((((((((.((((...................)))))))))))(((((.((....))...)))))..))). ( -47.01) >pau.2886 5475510 120 + 6537648 CAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCAUCCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCUUCCUUUAUCCGCGCGGACCGACGGCCCGCGCCAGCUCCGGCCGUCGGCC .......((...(((...((((((.(((((.....)))).).)))))).)))(((((.((((...................)))))))))(((((((.((....))...))))))))).. ( -50.41) >pel.1792 3328814 101 + 5888780 UCUUUUCCAGCAG----CCUUGCCACCCG------GCAUGGCG------CGUGGUCCUGCGCAUUCC-UCG-CUUUCAAUCGCGCCG-UCUUCGGCCGGCGCUGCCGCGGGCCAUCGCCC ............(----((.((((....)------))).)))(------(((((.(((((((.....-..)-)........((((((-.(....).))))))....)))))))).))).. ( -45.80) >consensus CAUUUUCCCUUCGUGUCCCUUCCCUCGCGCCA_CCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCUUUAUCCGCGCGGACCGUCGGCCCGCGCCAGCUCCGGCCGUCGGCC .....................((..((.(((....(((((((((....)))).....)))))...................((((((.((...)).)))))).......)))))..)).. (-26.25 = -26.23 + -0.02)

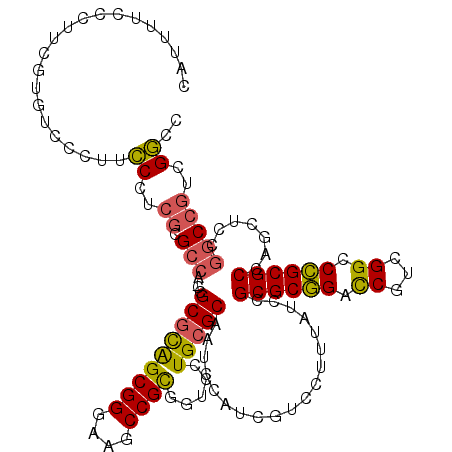
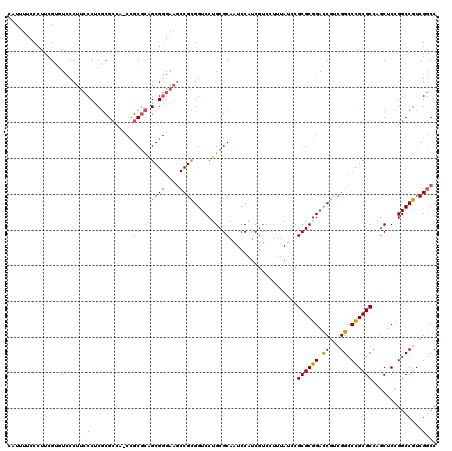
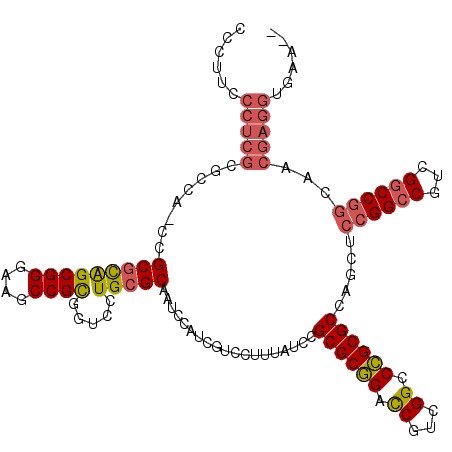
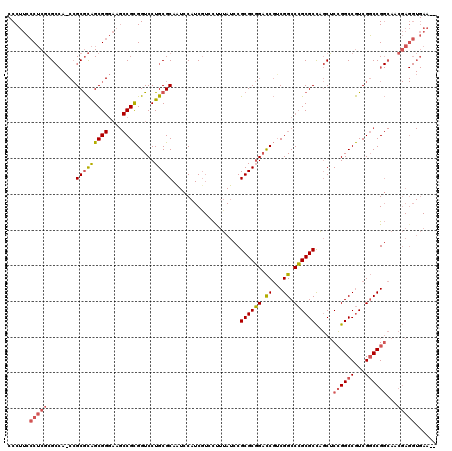
| Location | 5,203,547 – 5,203,667 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.88 |
| Mean single sequence MFE | -53.34 |
| Consensus MFE | -29.27 |
| Energy contribution | -31.03 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203547 120 + 6264404 CCCUUCCCUCGCGCCAACCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCCUUAUCCGCGCGGACCGUCGGCCCGCGCCAGCUCCGGCCGUCGGCCGGCAACGAGGUGAAAC ...(((((((((((.....)))).((((....))))(((((.((((...................)))))))))(((((((.(((((......)).))).)))))))...)))).))).. ( -58.91) >pau.2886 5475526 118 + 6537648 CCCUUCCCUCGCGCCAUCCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCUUCCUUUAUCCGCGCGGACCGACGGCCCGCGCCAGCUCCGGCCGUCGGCCGGCAACGAGGUGAA-- ....(((((((.(((.(((((((.((((....))))((...........))..............)))))))(((((((((.((....))...)))))))))..)))..))))).)).-- ( -58.80) >pel.1792 3328827 89 + 5888780 -CCUUGCCACCCG------GCAUGGCG------CGUGGUCCUGCGCAUUCC-UCG-CUUUCAAUCGCGCCG-UCUUCGGCCGGCGCUGCCGCGGGCCAUCGCCCG--------------- -...((((....)------))).((((------.((((.(((((((.....-..)-)........((((((-.(....).))))))....)))))))))))))..--------------- ( -42.30) >consensus CCCUUCCCUCGCGCCA_CCGCGCAGCGGGAAGCCGCGGUCCUGCGCAAUCCAUCGUCCUUUAUCCGCGCGGACCGUCGGCCCGCGCCAGCUCCGGCCGUCGGCCGGCAACGAGGUGAA__ ......(((((........(((((((((....)))).....)))))...................((((((.((...)).)))))).....((((((...))))))...)))))...... (-29.27 = -31.03 + 1.76)

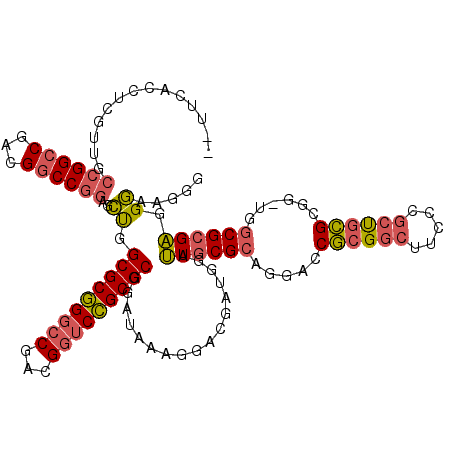
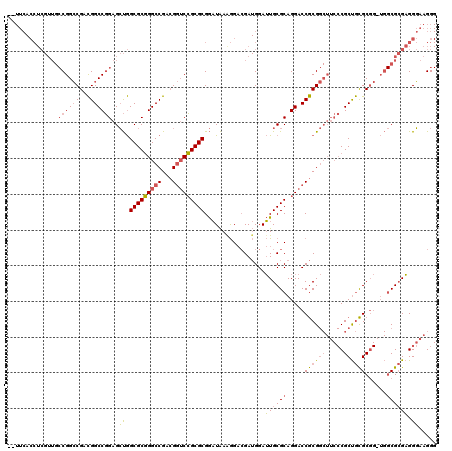
| Location | 5,203,547 – 5,203,667 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Mean single sequence MFE | -57.17 |
| Consensus MFE | -33.21 |
| Energy contribution | -34.33 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2750 5203547 120 - 6264404 GUUUCACCUCGUUGCCGGCCGACGGCCGGAGCUGGCGCGGGCCGACGGUCCGCGCGGAUAAGGGACGAUGGAUUGCGCAGGACCGCGGCUUCCCGCUGCGCGGUUGGCGCGAGGGAAGGG .((((.((((((.((((((((.((((((....))))((((((((.((((((((((((.((........))..)))))).))))))))))...))))..)))))))))))))))))))).. ( -67.80) >pau.2886 5475526 118 - 6537648 --UUCACCUCGUUGCCGGCCGACGGCCGGAGCUGGCGCGGGCCGUCGGUCCGCGCGGAUAAAGGAAGAUGGAUUGCGCAGGACCGCGGCUUCCCGCUGCGCGGAUGGCGCGAGGGAAGGG --(((.((((((.((((((((((((((.(.(....).).))))))))))((((((((.....(((((.....(((((......))))))))))..)))))))).)))))))))))))... ( -65.20) >pel.1792 3328827 89 - 5888780 ---------------CGGGCGAUGGCCCGCGGCAGCGCCGGCCGAAGA-CGGCGCGAUUGAAAG-CGA-GGAAUGCGCAGGACCACG------CGCCAUGC------CGGGUGGCAAGG- ---------------(((((....))))).(((.((((((........-))))))........(-((.-((...........)).))------)))).(((------(....))))...- ( -38.50) >consensus __UUCACCUCGUUGCCGGCCGACGGCCGGAGCUGGCGCGGGCCGACGGUCCGCGCGGAUAAAGGACGAUGGAUUGCGCAGGACCGCGGCUUCCCGCUGCGCGG_UGGCGCGAGGGAAGGG ..............((((((...))))))..((.(((((((((...))))))))).................((((((.....((((((.....))))))......)))))).))..... (-33.21 = -34.33 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:56 2007