
| Sequence ID | pao.731 |
|---|---|
| Location | 1,292,452 – 1,292,570 |
| Length | 118 |
| Max. P | 0.987600 |

| Location | 1,292,452 – 1,292,558 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.21 |
| Mean single sequence MFE | -52.10 |
| Consensus MFE | -23.84 |
| Energy contribution | -28.62 |
| Covariance contribution | 4.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.731 1292452 106 + 6264404 AGCGCC------------GGGCGCGCAUGGACAGCGCCACCCGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGAGCCCGCGUCGCUCGCGGGG--UUAGAG ((((((------------(((.((((.......))))..))))))))).(((((.((((((((((.....))))))...))))..))))).((.((((((.....)))))).--)).... ( -58.70) >pau.2299 2181088 106 - 6537648 AGCGCC------------GGGCGCGCAUGGACAGCGCCACCUGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGACCCCGCGUCGCUCGCGGGG--UUAGAG ((((((------------(((.((((.......))))..))))))))).(((((.((((((((((.....))))))...))))..))))).(((((((((.....)))))))--)).... ( -61.00) >pfo.2488 1663725 107 - 6438405 AGCGCCUACGGGCCAAUCGGCCGAGCAAAUACAGGUCUUCAGAUUG-----------UGUGCGACCUUAUGUCGCAGUAUAGAAAGGUCUUAA--CCGCACAGGUGGUAGGGGCUUAGAG (((.(((((.((((....))))..((..((((.((((....)))).-----------..((((((.....)))))))))).....(((....)--)))).......))))).)))..... ( -36.60) >consensus AGCGCC____________GGGCGCGCAUGGACAGCGCCACCCGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGA_CCCGCGUCGCUCGCGGGG__UUAGAG .(((((.............)))))........((((((....)))))).(((((.((((((((((.....))))))...))))..)))))....((((((.....))))))......... (-23.84 = -28.62 + 4.78)

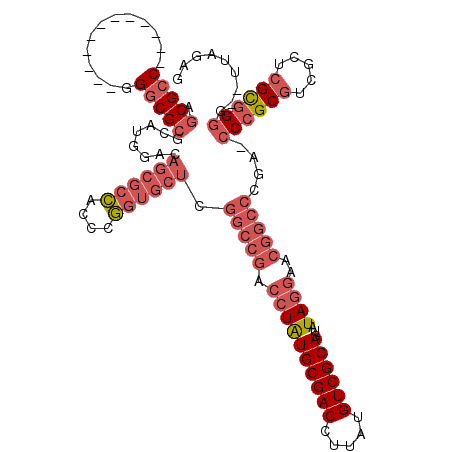
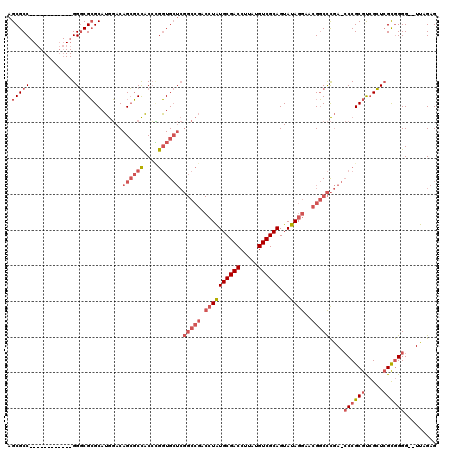
| Location | 1,292,452 – 1,292,558 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.21 |
| Mean single sequence MFE | -48.60 |
| Consensus MFE | -23.38 |
| Energy contribution | -27.72 |
| Covariance contribution | 4.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.731 1292452 106 - 6264404 CUCUAA--CCCCGCGAGCGACGCGGGCUCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCGGGUGGCGCUGUCCAUGCGCGCCC------------GGCGCU ......--.((((((.....))))))....(((((..((((...((((((.....)))))))))).))))).(((.(((((((.(((.......))))))))------------)).))) ( -58.20) >pau.2299 2181088 106 + 6537648 CUCUAA--CCCCGCGAGCGACGCGGGGUCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCAGGUGGCGCUGUCCAUGCGCGCCC------------GGCGCU .....(--(((((((.....))))))))..(((((..((((...((((((.....)))))))))).))))).(((.((.((((.(((.......))))))).------------)).))) ( -56.80) >pfo.2488 1663725 107 + 6438405 CUCUAAGCCCCUACCACCUGUGCGG--UUAAGACCUUUCUAUACUGCGACAUAAGGUCGCACA-----------CAAUCUGAAGACCUGUAUUUGCUCGGCCGAUUGGCCCGUAGGCGCU .....(((.(((((.....((((((--((.(((....)))....((((((.....))))))..-----------.........)))).))))......((((....)))).))))).))) ( -30.80) >consensus CUCUAA__CCCCGCGAGCGACGCGGG_UCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCAGGUGGCGCUGUCCAUGCGCGCCC____________GGCGCU .........((((((.....))))))....(((((..((((...((((((.....)))))))))).))))).(((........((((.......))))(((.............)))))) (-23.38 = -27.72 + 4.34)

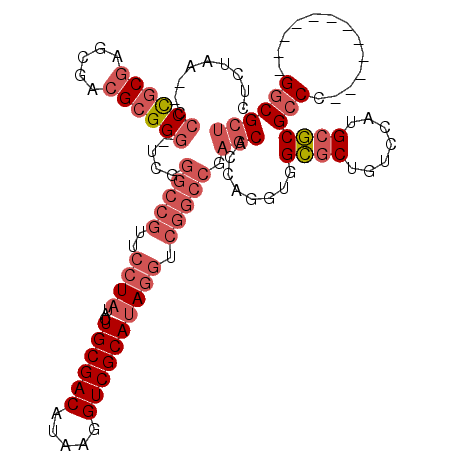
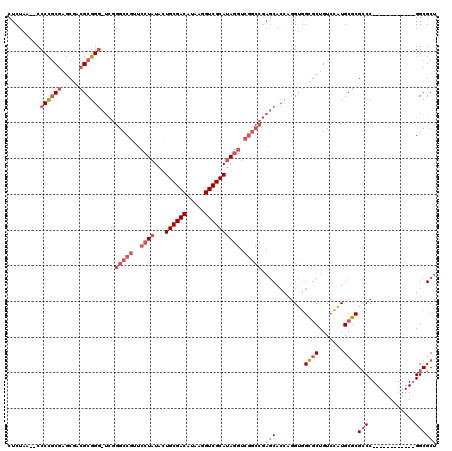
| Location | 1,292,458 – 1,292,570 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.14 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -25.28 |
| Energy contribution | -29.40 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
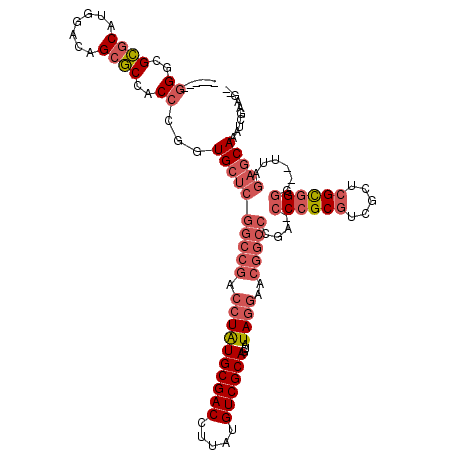
>pao.731 1292458 112 + 6264404 ------GGGCGCGCAUGGACAGCGCCACCCGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGAGCCCGCGUCGCUCGCGGGG--UUAGAGCAAAUCGAAGCU ------.(((((.........)))))...(((((((((((((.((((((((((.....))))))...))))..))))).((.((((((.....)))))).--)).)))))..)))..... ( -53.60) >pau.2299 2181094 110 - 6537648 ------GGGCGCGCAUGGACAGCGCCACCUGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGACCCCGCGUCGCUCGCGGGG--UUAGAGCAAAUCGAAG-- ------.(((((.........)))))......((((((((((.((((((((((.....))))))...))))..))))).(((((((((.....)))))))--)).)))))........-- ( -57.10) >pfo.2488 1663737 102 - 6438405 CCAAUCGGCCGAGCAAAUACAGGUCUUCAGAUUG-----------UGUGCGACCUUAUGUCGCAGUAUAGAAAGGUCUUAA--CCGCACAGGUGGUAGGGGCUUAGAGCAAAUCG----- ......((((...........))))....(((((-----------(.((((((.....))))))........((((((..(--((((....)))))..))))))...)).)))).----- ( -29.10) >consensus ______GGGCGCGCAUGGACAGCGCCACCCGGUGCUCGGCCGACCUAUGCGACCUUAUGUCGCAGUAUAGGAACGGCCCGA_CCCGCGUCGCUCGCGGGG__UUAGAGCAAAUCGAAG__ ......((..((((.......))))..))...((((((((((.((((((((((.....))))))...))))..)))))....((((((.....))))))......))))).......... (-25.28 = -29.40 + 4.12)

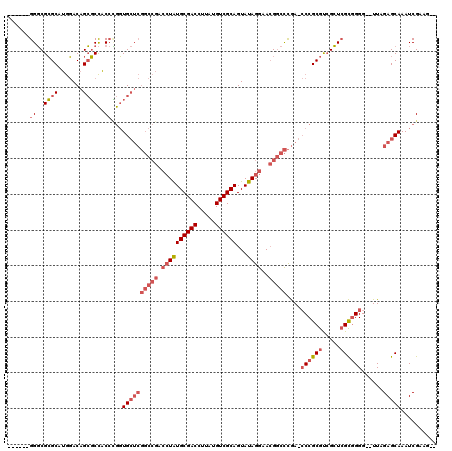
| Location | 1,292,458 – 1,292,570 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.14 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -25.83 |
| Energy contribution | -30.17 |
| Covariance contribution | 4.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
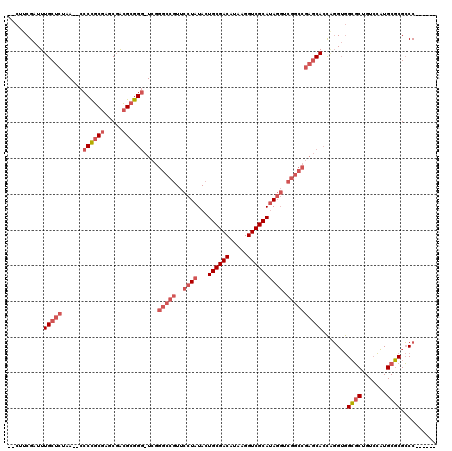
>pao.731 1292458 112 - 6264404 AGCUUCGAUUUGCUCUAA--CCCCGCGAGCGACGCGGGCUCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCGGGUGGCGCUGUCCAUGCGCGCCC------ .((.(((.(((((.....--....)))))))).))((((((((.(((..((((...((((((.....)))))))))).))))))))).))(((((.(((.......))))))))------ ( -55.00) >pau.2299 2181094 110 + 6537648 --CUUCGAUUUGCUCUAA--CCCCGCGAGCGACGCGGGGUCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCAGGUGGCGCUGUCCAUGCGCGCCC------ --........(((((..(--(((((((.....))))))))..(((((..((((...((((((.....)))))))))).))))))))))...((((.(((.......))))))).------ ( -56.20) >pfo.2488 1663737 102 + 6438405 -----CGAUUUGCUCUAAGCCCCUACCACCUGUGCGG--UUAAGACCUUUCUAUACUGCGACAUAAGGUCGCACA-----------CAAUCUGAAGACCUGUAUUUGCUCGGCCGAUUGG -----(((((.(((...(((...(((....(((((((--(..(((....)))..))))).)))..(((((...((-----------.....))..))))))))...))).))).))))). ( -23.30) >consensus __CUUCGAUUUGCUCUAA__CCCCGCGAGCGACGCGGG_UCGGGCCGUUCCUAUACUGCGACAUAAGGUCGCAUAGGUCGGCCGAGCACCAGGUGGCGCUGUCCAUGCGCGCCC______ ..........(((((......((((((.....))))))....(((((..((((...((((((.....)))))))))).)))))))))).......((((.......)))).......... (-25.83 = -30.17 + 4.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:44 2007