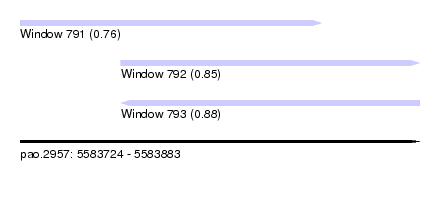
| Sequence ID | pao.2957 |
|---|---|
| Location | 5,583,724 – 5,583,883 |
| Length | 159 |
| Max. P | 0.877452 |

| Location | 5,583,724 – 5,583,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.89 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2957 5583724 120 + 6264404 AAGGCAGGCAUGGCGCGGACAUCUUGUUCCCUACGCGGGUUGCUGCGGUCCGCGCGAGGCGGAACCGGACGCUAGCCCGAUCAGGUUCAACGGGAUCCGGAACUAUCCGAUCUCAGCCCC ..(((..((.(.((((((((.....((.(((.....)))..))....)))))))).).)).(((((.((((......)).)).)))))...((((((.(((....))))))))).))).. ( -50.00) >ppk.2856 5596227 79 + 6181863 AAGGCGAGCA----------AUCUUGUUCCCUACGCAGG------------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGCACCUCUGCGAUCUCAGCC-U .(((.(((((----------....))))))))....(((------------------------------(...(((((....)))))....((((((.(((....))))))))).)))-) ( -27.70) >pst.2969 5648630 76 + 6397126 -AGACGAGAA----------AUCUUGUUCCCUACGCAGG------------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCG-GUUUAACCGAUCUCAGCC-- -.((((((..----------..)))))).........((------------------------------(...(((((....)))))....((((((.(-(.....)))))))).)))-- ( -22.50) >pf5.316 6447133 76 - 7074893 -UGACGAGAA----------AUCUUGUUCCCUACGCAGG------------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCG-GAUUAUCCGAUCUCAGCC-- -....((((.----------((((((((.(((...((((------------------------------......))))...)))...)))))))).((-(.....))).))))....-- ( -22.60) >pfo.264 5868437 77 - 6438405 -UGACGAGAA----------AUCUUGUUCCCUACGCAGG------------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGAAAUUAUUCGAUCUCAGCC-- -....((((.----------((((((((.(((...((((------------------------------......))))...)))...)))))))).((((....)))).))))....-- ( -20.80) >pel.2860 5282419 79 + 5888780 AAUGAGAGCA----------AUCUUGUUCCCUACGCAGG------------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGCAACUUUGCGAUCUCAGCC-U ((..(((...----------.)))..))........(((------------------------------(...(((((....)))))....((((((.(((....))))))))).)))-) ( -24.60) >consensus _AGACGAGAA__________AUCUUGUUCCCUACGCAGG______________________________CGCUAACCUGAUCAGGUUCAACGGGAUCCG_AACUAUCCGAUCUCAGCC__ ..((((((..............)))))).(((....)))...............................((((((((....)))))....((((((.(((....))))))))))))... (-16.08 = -15.89 + -0.19)

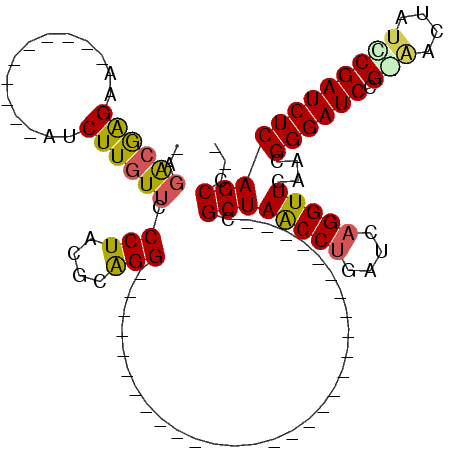
| Location | 5,583,764 – 5,583,883 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2957 5583764 119 + 6264404 UGCUGCGGUCCGCGCGAGGCGGAACCGGACGCUAGCCCGAUCAGGUUCAACGGGAUCCGGAACUAUCCGAUCUCAGCCCCUCGUA-UAGGGCACCCCGACAAGAACCCCGGCAGUCUAAU .(((((((((.(.((..((((........)))).))).)))).(((((...((((((.(((....))))))))).((((......-..))))..........)))))...)))))..... ( -44.60) >ppk.2856 5596256 90 + 6181863 -----------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGCACCUCUGCGAUCUCAGCC-UUUGCUUCAAGGCACCCCGACAAGAACAUGCGCAGUCUAGA -----------------------------..(((..(((..((.((((...((((((.(((....))))))))).(((-((......)))))..........)))).))..)))..))). ( -27.60) >pss.328 5478709 88 - 6093698 -----------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCG-GUUUAACCGAUCUCAGCC--UCAUAGCAAGGCACCCCGACAAGAACGCCGCCAGUCUAUG -----------------------------.((((((((....)))))....((((((.(-(.....))))))))))).--.(((((...(((....((.......))..)))...))))) ( -22.10) >pf5.316 6447161 88 - 7074893 -----------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCG-GAUUAUCCGAUCUCAGCC--UCAUAGCAAGGCACCCCGACAAGAACCGGGCCAGUCUAGA -----------------------------..(((..(((..(.(((((...((((((.(-(.....)))))))).(((--(.......))))..........))))).)..)))..))). ( -30.40) >pfo.264 5868465 89 - 6438405 -----------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGAAAUUAUUCGAUCUCAGCC--UCAUAGCAAGGCACCCCGACAAGAACCCGGCCAGUCUAGA -----------------------------..(((..(((..(.(((((...((((((.(((....))))))))).(((--(.......))))..........))))).)..)))..))). ( -28.60) >pel.2860 5282448 90 + 5888780 -----------------------------CGCUAACCUGAUCAGGUUCAACGGGAUCCGCAACUUUGCGAUCUCAGCC-UUUGCUUCAAGGCACCCCGACAAGAACAUGCGCAGUCUAGA -----------------------------..(((..(((..((.((((...((((((.(((....))))))))).(((-((......)))))..........)))).))..)))..))). ( -27.70) >consensus _____________________________CGCUAACCUGAUCAGGUUCAACGGGAUCCG_AACUAUCCGAUCUCAGCC__UCAUAGCAAGGCACCCCGACAAGAACCCGGCCAGUCUAGA ...............................(((..(((.....((((...((((((.(((....))))))))).(((...........)))..........)))).....)))..))). (-17.05 = -17.67 + 0.61)

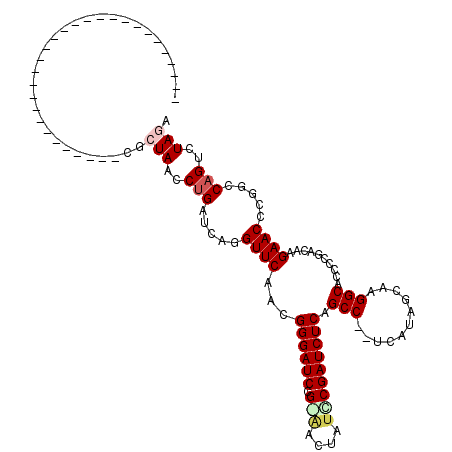

| Location | 5,583,764 – 5,583,883 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2957 5583764 119 - 6264404 AUUAGACUGCCGGGGUUCUUGUCGGGGUGCCCUA-UACGAGGGGCUGAGAUCGGAUAGUUCCGGAUCCCGUUGAACCUGAUCGGGCUAGCGUCCGGUUCCGCCUCGCGCGGACCGCAGCA ....(.((((.(((((((....(((((((((((.-.....))))).....(((((....)))))))))))..)))))..(((((((....)))))))(((((.....)))))))))))). ( -53.60) >ppk.2856 5596256 90 - 6181863 UCUAGACUGCGCAUGUUCUUGUCGGGGUGCCUUGAAGCAAA-GGCUGAGAUCGCAGAGGUGCGGAUCCCGUUGAACCUGAUCAGGUUAGCG----------------------------- .((((.(((..((.((((....(((((((((((......))-))).....(((((....)))))))))))..)))).))..))).))))..----------------------------- ( -31.00) >pss.328 5478709 88 + 6093698 CAUAGACUGGCGGCGUUCUUGUCGGGGUGCCUUGCUAUGA--GGCUGAGAUCGGUUAAAC-CGGAUCCCGUUGAACCUGAUCAGGUUAGCG----------------------------- ......(((((((.((((....(((((((((((.....))--))).....((((.....)-)))))))))..))))))).)))).......----------------------------- ( -31.50) >pf5.316 6447161 88 + 7074893 UCUAGACUGGCCCGGUUCUUGUCGGGGUGCCUUGCUAUGA--GGCUGAGAUCGGAUAAUC-CGGAUCCCGUUGAACCUGAUCAGGUUAGCG----------------------------- .((((.(((..(.(((((....(((((((((((.....))--))).....((((.....)-)))))))))..))))).)..))).))))..----------------------------- ( -34.60) >pfo.264 5868465 89 + 6438405 UCUAGACUGGCCGGGUUCUUGUCGGGGUGCCUUGCUAUGA--GGCUGAGAUCGAAUAAUUUCGGAUCCCGUUGAACCUGAUCAGGUUAGCG----------------------------- .((((.(((..(((((((....(((((((((((.....))--))).....(((((....)))))))))))..)))))))..))).))))..----------------------------- ( -37.50) >pel.2860 5282448 90 - 5888780 UCUAGACUGCGCAUGUUCUUGUCGGGGUGCCUUGAAGCAAA-GGCUGAGAUCGCAAAGUUGCGGAUCCCGUUGAACCUGAUCAGGUUAGCG----------------------------- .((((.(((..((.((((....(((((((((((......))-))).....(((((....)))))))))))..)))).))..))).))))..----------------------------- ( -31.10) >consensus UCUAGACUGCCCGGGUUCUUGUCGGGGUGCCUUGCUACGA__GGCUGAGAUCGGAUAAUU_CGGAUCCCGUUGAACCUGAUCAGGUUAGCG_____________________________ .((((.(((..((.((((....(((((((((...........))).....(((((....)))))))))))..)))).))..))).))))............................... (-22.50 = -23.00 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:35 2007