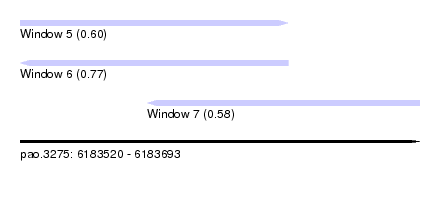
| Sequence ID | pao.3275 |
|---|---|
| Location | 6,183,520 – 6,183,693 |
| Length | 173 |
| Max. P | 0.768746 |

| Location | 6,183,520 – 6,183,636 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -26.74 |
| Energy contribution | -25.47 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
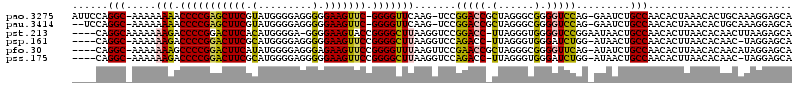
>pao.3275 6183520 116 + 6264404 AUUCCAGGC-AAAAAAAACCCCGAGCUUCGUAUGGGGAGGGGGAAGUUC-GGGGUUCAAG-UCCGGACCGCUAGGGCGGGGUCCAG-GAAUCUGCCAACACUAAACACUGCAAAGGAGCA .((((.(((-(.....(((((((((((((.(.........).)))))))-))))))....-((((((((.(......).))))).)-))...))))..((........))....)))).. ( -43.10) >pau.3414 6456433 114 + 6537648 --UCCAGGC-AAAAAAAACCCCGAGCUUCGUAUGGGGAGGGGGAAGUUC-GGGGUUCAAG-UCCGGACCGCUAGGGCGGGGUCCAG-GAAUCUGCCAACACUAAACACUGCAAAGGAGCA --....(((-(.....(((((((((((((.(.........).)))))))-))))))....-((((((((.(......).))))).)-))...)))).............((......)). ( -42.80) >pst.213 373001 114 + 6397126 ----CAGGCAAAAAAAGACCCCGGACUUCACAUGGGGA-GGGGAAGUACCGGGGCUUAAGGUCCGGACC-UUAGGGUGGGGUCCGGAAUAACUGCCAACACUUAACACAACUUAAGAGCA ----..((((....(((.(((((((((((.(.......-.).))))).)))))))))....((((((((-((.....)))))))))).....))))....(((((......))))).... ( -45.70) >psp.161 299555 112 + 5928787 ----CAGGC-AAAAAAGACCCCGGACUUCGCAUGGGGAGGGGGAAGUUCCGGGGCUUAAGGUCCAGACC-UUAGGGUGGGAUCUGG-AUAACUGCCAACACUUAACACAAC-UAGGAGCA ----..(((-(...(((.(((((((((((.(.........).)))).))))))))))...(((((((((-(......))).)))))-))...))))....((((.......-)))).... ( -41.80) >pfo.30 60766 114 + 6438405 ----CAGGC-AAAAAAAGCCCCGGACUUCAUAUGGGGAGGGAGAAGUUCCGGGGUUUAAGUUCCGAACCGCUAGGGCGGGGUUCAG-AUAUCUGCCAACACUUAACACAACAUAGGAGCA ----.....-.....((((((((((((((.............)))).))))))))))..(((((...((((....))))((..(((-....)))))..................))))). ( -39.02) >pss.175 287452 112 + 6093698 ----CAGGC-AAAAAAGACCCCGGACUUCGCAUGGGGAGGGGGAAGUUCCGGGGCUUAAGGUCCAGACC-UUAGGGUGGGAUCUGG-AUAACUGCCAACACUUAACACAAC-UAGGAGCA ----..(((-(...(((.(((((((((((.(.........).)))).))))))))))...(((((((((-(......))).)))))-))...))))....((((.......-)))).... ( -41.80) >consensus ____CAGGC_AAAAAAAACCCCGGACUUCGCAUGGGGAGGGGGAAGUUCCGGGGCUUAAGGUCCGGACC_CUAGGGCGGGGUCCAG_AUAACUGCCAACACUUAACACAACAUAGGAGCA ......(((.....(((.(((((((((((.(.........).))))))).))))))).......(((((.(......).))))).........)))........................ (-26.74 = -25.47 + -1.28)
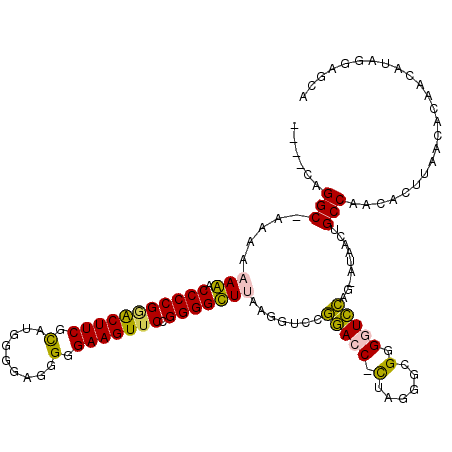
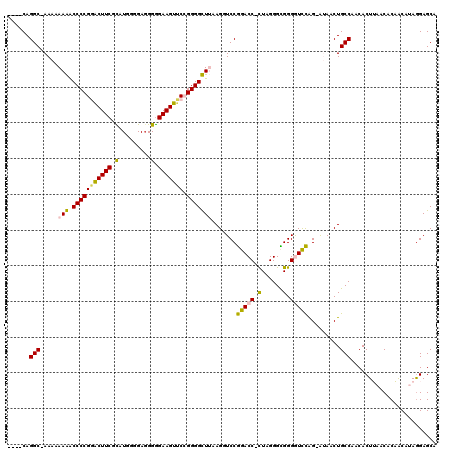
| Location | 6,183,520 – 6,183,636 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -30.00 |
| Energy contribution | -28.54 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3275 6183520 116 - 6264404 UGCUCCUUUGCAGUGUUUAGUGUUGGCAGAUUC-CUGGACCCCGCCCUAGCGGUCCGGA-CUUGAACCCC-GAACUUCCCCCUCCCCAUACGAAGCUCGGGGUUUUUUUU-GCCUGGAAU (((......)))............((((((...-(((((((.(......).))))))).-...(((((((-((.((((.............)))).)))))))))..)))-)))...... ( -39.72) >pau.3414 6456433 114 - 6537648 UGCUCCUUUGCAGUGUUUAGUGUUGGCAGAUUC-CUGGACCCCGCCCUAGCGGUCCGGA-CUUGAACCCC-GAACUUCCCCCUCCCCAUACGAAGCUCGGGGUUUUUUUU-GCCUGGA-- (((......)))............((((((...-(((((((.(......).))))))).-...(((((((-((.((((.............)))).)))))))))..)))-)))....-- ( -39.72) >pst.213 373001 114 - 6397126 UGCUCUUAAGUUGUGUUAAGUGUUGGCAGUUAUUCCGGACCCCACCCUAA-GGUCCGGACCUUAAGCCCCGGUACUUCCCC-UCCCCAUGUGAAGUCCGGGGUCUUUUUUUGCCUG---- .((.(((((......))))).)).(((((....((((((((.........-))))))))......(((((((.(((((.(.-.......).))))))))))))......)))))..---- ( -41.90) >psp.161 299555 112 - 5928787 UGCUCCUA-GUUGUGUUAAGUGUUGGCAGUUAU-CCAGAUCCCACCCUAA-GGUCUGGACCUUAAGCCCCGGAACUUCCCCCUCCCCAUGCGAAGUCCGGGGUCUUUUUU-GCCUG---- ........-...............(((((...(-(((((((.........-))))))))......((((((((.((((.............)))))))))))).....))-)))..---- ( -34.72) >pfo.30 60766 114 - 6438405 UGCUCCUAUGUUGUGUUAAGUGUUGGCAGAUAU-CUGAACCCCGCCCUAGCGGUUCGGAACUUAAACCCCGGAACUUCUCCCUCCCCAUAUGAAGUCCGGGGCUUUUUUU-GCCUG---- ........................((((((..(-(((((((.(......).)))))))).......(((((((.((((.............))))))))))).....)))-)))..---- ( -37.12) >pss.175 287452 112 - 6093698 UGCUCCUA-GUUGUGUUAAGUGUUGGCAGUUAU-CCAGAUCCCACCCUAA-GGUCUGGACCUUAAGCCCCGGAACUUCCCCCUCCCCAUGCGAAGUCCGGGGUCUUUUUU-GCCUG---- ........-...............(((((...(-(((((((.........-))))))))......((((((((.((((.............)))))))))))).....))-)))..---- ( -34.72) >consensus UGCUCCUAUGUUGUGUUAAGUGUUGGCAGAUAU_CCGGACCCCACCCUAA_GGUCCGGACCUUAAACCCCGGAACUUCCCCCUCCCCAUACGAAGUCCGGGGUCUUUUUU_GCCUG____ ........................(((.......(((((((..........))))))).....(((((((((.(((((.............))))))))))).))).....)))...... (-30.00 = -28.54 + -1.47)
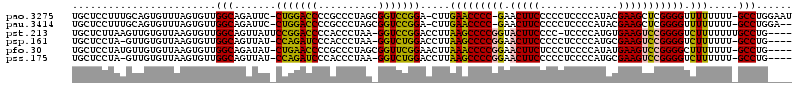
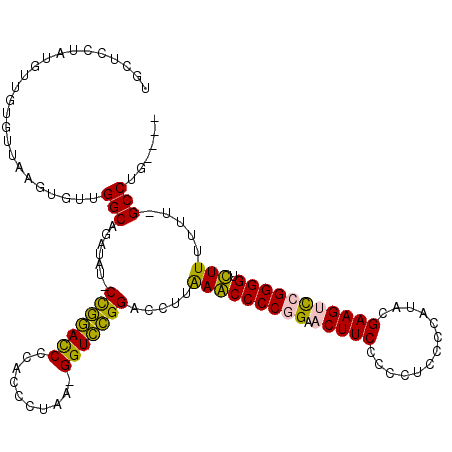
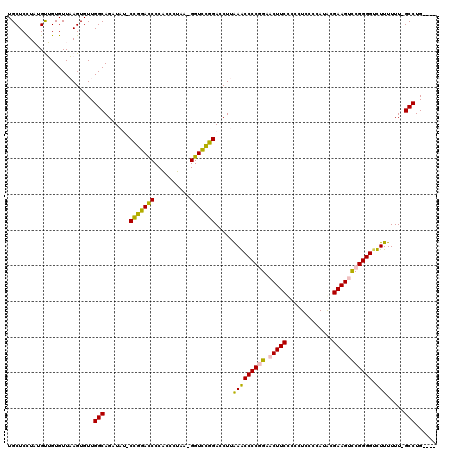
| Location | 6,183,575 – 6,183,693 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -23.16 |
| Energy contribution | -21.50 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3275 6183575 118 - 6264404 GAACUUUCCCGUUUUGCCGACAGUCAGAACACACGAAUGGCUUCUUGAGCCCUUCGAUGCUCCUUUGCAGUGUUUAGUGUUGGCAGAUUC-CUGGACCCCGCCCUAGCGGUCCGGA-CUU ............((((((((((.(..((((((.((((.(((((...))))).)))).(((......)))))))))).))))))))))...-(((((((.(......).))))))).-... ( -45.80) >pau.3414 6456486 118 - 6537648 GAACUUUCCCGUUUUGCCGACAGUCAGAACACACGAAUGGCUUCUUGAGCCCUUCGAUGCUCCUUUGCAGUGUUUAGUGUUGGCAGAUUC-CUGGACCCCGCCCUAGCGGUCCGGA-CUU ............((((((((((.(..((((((.((((.(((((...))))).)))).(((......)))))))))).))))))))))...-(((((((.(......).))))))).-... ( -45.80) >pst.213 373053 117 - 6397126 GAACUUAACCGUUUGGACAGGAGUCAGAGGUUCCGAUUGAUUGA-AAAAUCCAUCG-UGCUCUUAAGUUGUGUUAAGUGUUGGCAGUUAUUCCGGACCCCACCCUAA-GGUCCGGACCUU ..(((((((((....(((....)))((((....((((.((((..-..)))).))))-..)))).....)).)))))))............((((((((.........-)))))))).... ( -33.00) >psp.161 299607 115 - 5928787 GAACUUAACCGUUUGGAUAGGAGUCAGAGGUCUCGAUUGGUUGA-AAAAUCGAUCG-UGCUCCUA-GUUGUGUUAAGUGUUGGCAGUUAU-CCAGAUCCCACCCUAA-GGUCUGGACCUU ..(((((((((......(((((((..((....))((((((((..-..)))))))).-.)))))))-..)).)))))))...........(-(((((((.........-)))))))).... ( -35.30) >ppk.74 6051361 118 + 6181863 GAACUAAACCGGUGAAGCGGAACUCAGAGUAACUGAAUGGCUGGUGAAGCCCUUCGAUGCUCCUAUGUUGUGUUAAGUGUUGGCAGAUAU-CUGGACCCCGCCCUAGCGGUCCGGA-CUU ........(((..(.((((.(((...(((((..((((.((((.....)))).)))).)))))....))).))))...)..)))......(-(((((((.(......).))))))))-... ( -35.00) >pel.46 5814211 118 + 5888780 GAACUAAAUCGGUGAAGCGCUACUCAGAGGUACUGAAUGGCUGGUGAAGCCUUUCGAUGCUCCUAUGUUGUGUUAAGUGUUGGCAGAUAU-CUGGACCCCGCCCUAGCGGUCCGGA-CUU .......(((.((..((((((((..((..(((.((((.((((.....)))).)))).)))..))..)).......)))))).)).))).(-(((((((.(......).))))))))-... ( -35.71) >consensus GAACUUAACCGUUUAGACGGCAGUCAGAGGACCCGAAUGGCUGAUAAAGCCCUUCGAUGCUCCUAUGUUGUGUUAAGUGUUGGCAGAUAU_CUGGACCCCGCCCUAGCGGUCCGGA_CUU ......................(((((......((((.((((.....)))).))))..((......))...........))))).......(((((((.(......).)))))))..... (-23.16 = -21.50 + -1.66)
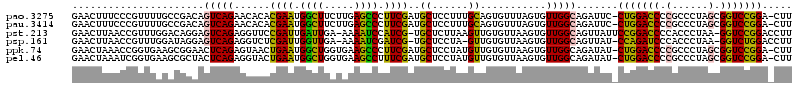
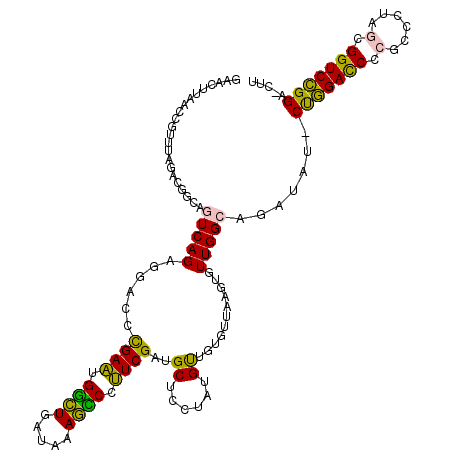
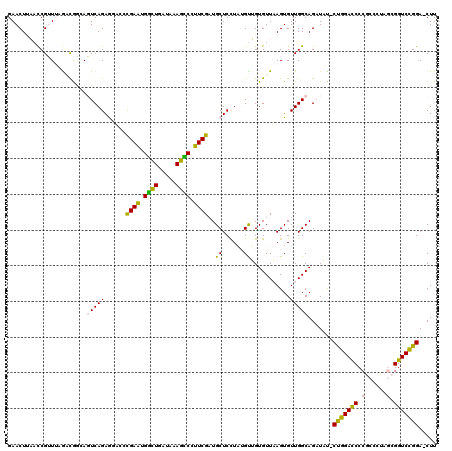
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:20 2007