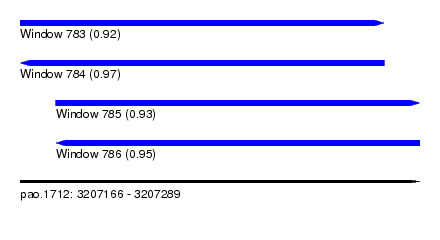
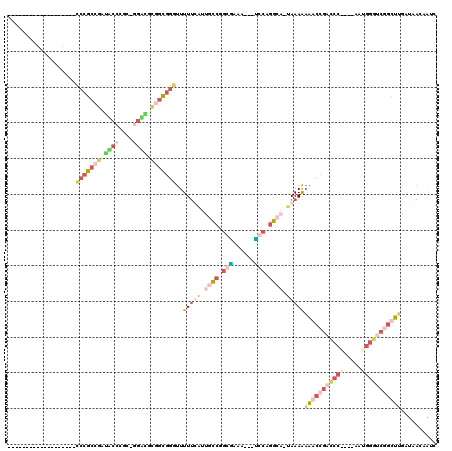
| Sequence ID | pao.1712 |
|---|---|
| Location | 3,207,166 – 3,207,289 |
| Length | 123 |
| Max. P | 0.971767 |

| Location | 3,207,166 – 3,207,278 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 43.02 |
| Mean single sequence MFE | -54.10 |
| Consensus MFE | -24.70 |
| Energy contribution | -29.32 |
| Covariance contribution | 4.62 |
| Combinations/Pair | 1.66 |
| Mean z-score | -5.48 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921885 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.1712 3207166 112 + 6264404 UAGGUCUCACGACCUGUUGAAAACCGGUCCCUCG----GGGCCGGUUUUUUUA-UGCCUGCG---UUUCGCCGGGCAUGAAAAAACCCGCGUCCCUUGCGGGUACGCGGGCCUCUGCCUC ((((((....))))))..((((((((((((....----))))))))))))(((-((((((((---...)).))))))))).....((((((((((....))).))))))).......... ( -57.90) >pau.1282 4175432 112 - 6537648 CGCAAUUUACUCGAACAUUUCCCGCUUGGCAAAU----AUUUUUUUAACUUUU-UCAUCGGC---UUAGGUCGUCGGAGGAAAGUCCGCGGGGUGCCUUGGUAGACUGCCGGGCACGAAG ..........((........(((((..((((((.----.....)))...((((-((..((((---....))))..))))))..))).)))))((((((.(((.....))))))))))).. ( -32.40) >pf5.2596 4971749 112 + 7074893 UAGUCCCUCGGGAUUGUUAUCGAGCCGAUCCAUUUUUUGGAUCGGCUUUUUUAUUGCCCGGAGUUCUUCACCGGCAAUAAAAAACCCGCCGCUACCAGCAGGUAUCGGCGGG-------- ((((((....)))))).....(((((((((((.....)))))))))))((((((((((.((.(.....).))))))))))))..(((((((.((((....)))).)))))))-------- ( -58.20) >pfo.2361 4537202 109 + 6438405 UAGUCCUUCGGGAUUGUUAUCAAGCCGACCCGUUUUUCGGGUCGGCUUUUUUG-UGCCUGCAGUUUUCUGCGGGCAAUAAAAAACCCGCCACGGC--GCAAGCCUCGGCGGG-------- ((((((....)))))).....(((((((((((.....)))))))))))(((((-(((((((((....)))))))).))))))..((((((..(((--....)))..))))))-------- ( -67.90) >consensus UAGUCCUUACGGAUUGUUAUCAAGCCGACCCAUU____GGGUCGGCUUUUUUA_UGCCCGCA___UUUCGCCGGCAAUAAAAAACCCGCCGCGACC_GCAGGUAUCGGCGGG________ ((((((....)))))).......(((((((((.....)))))))))(((((((.((((.((((....)))).)))).)))))))(((((((.((((....)))).)))))))........ (-24.70 = -29.32 + 4.62)

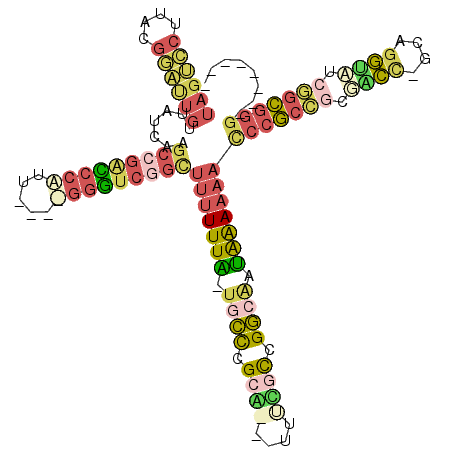
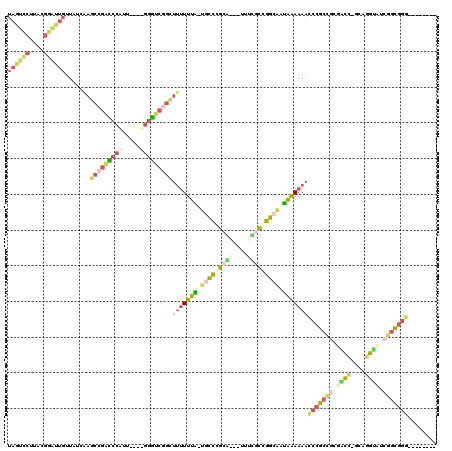
| Location | 3,207,166 – 3,207,278 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 43.02 |
| Mean single sequence MFE | -49.12 |
| Consensus MFE | -17.90 |
| Energy contribution | -23.77 |
| Covariance contribution | 5.88 |
| Combinations/Pair | 1.54 |
| Mean z-score | -4.62 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971767 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.1712 3207166 112 - 6264404 GAGGCAGAGGCCCGCGUACCCGCAAGGGACGCGGGUUUUUUCAUGCCCGGCGAAA---CGCAGGCA-UAAAAAAACCGGCCC----CGAGGGACCGGUUUUCAACAGGUCGUGAGACCUA ..((.((((((((((((.(((....)))))))))))))))))(((((..(((...---))).))))-)...(((((((((((----...))).))))))))....(((((....))))). ( -61.50) >pau.1282 4175432 112 + 6537648 CUUCGUGCCCGGCAGUCUACCAAGGCACCCCGCGGACUUUCCUCCGACGACCUAA---GCCGAUGA-AAAAGUUAAAAAAAU----AUUUGCCAAGCGGGAAAUGUUCGAGUAAAUUGCG .((((....((((.((((....))))....((((((......)))).))......---))))....-............(((----((((.((....)).)))))))))))......... ( -25.00) >pf5.2596 4971749 112 - 7074893 --------CCCGCCGAUACCUGCUGGUAGCGGCGGGUUUUUUAUUGCCGGUGAAGAACUCCGGGCAAUAAAAAAGCCGAUCCAAAAAAUGGAUCGGCUCGAUAACAAUCCCGAGGGACUA --------(((((((.((((....)))).)))))))((((((((((((.(.((.....))).))))))))))))(((((((((.....)))))))))..........(((....)))... ( -55.90) >pfo.2361 4537202 109 - 6438405 --------CCCGCCGAGGCUUGC--GCCGUGGCGGGUUUUUUAUUGCCCGCAGAAAACUGCAGGCA-CAAAAAAGCCGACCCGAAAAACGGGUCGGCUUGAUAACAAUCCCGAAGGACUA --------.((((((.(((....--))).))))))(((((((..((((.((((....)))).))))-.....(((((((((((.....)))))))))))............))))))).. ( -54.10) >consensus ________CCCGCCGAUACCCGC_GGACGCGGCGGGUUUUUCAUUGCCGGCGAAA___UCCAGGCA_UAAAAAAACCGACCC____AAUGGGUCGGCUUGAUAACAAUCCCGAAGGACUA ........(((((((.((((....)))).)))))))..(((((.((((.(((......))).)))).)))))(((((((((((.....)))))))))))......(((((....))))). (-17.90 = -23.77 + 5.88)

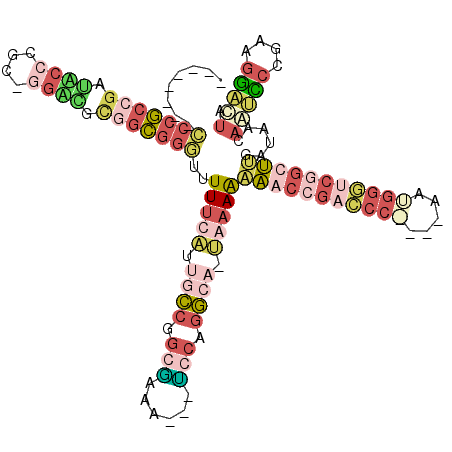
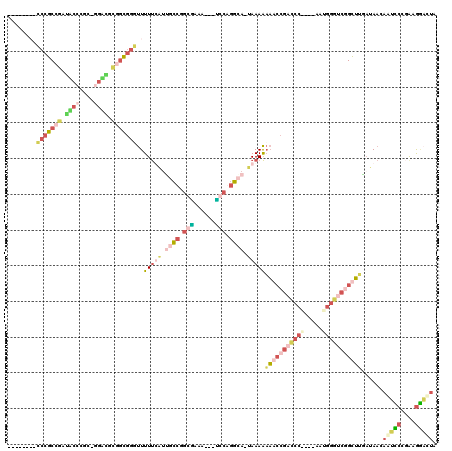
| Location | 3,207,177 – 3,207,289 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 40.17 |
| Mean single sequence MFE | -50.03 |
| Consensus MFE | -22.60 |
| Energy contribution | -25.60 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.69 |
| Mean z-score | -4.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934057 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.1712 3207177 112 + 6264404 ACCUGUUGAAAACCGGUCCCUCG----GGGCCGGUUUUUUUA-UGCCUGCG---UUUCGCCGGGCAUGAAAAAACCCGCGUCCCUUGCGGGUACGCGGGCCUCUGCCUCGACGCUCUCCG ....(((((..((((((((....----))))))))(((((((-((((((((---...)).))))))))))))).((((((((((....))).)))))))........)))))........ ( -55.10) >pau.1282 4175443 110 - 6537648 CGAACAUUUCCCGCUUGGCAAAU----AUUUUUUUAACUUUU-UCAUCGGC---UUAGGUCGUCGGAGGAAAGUCCGCGGGGUGCCUUGGUAGACUGCCGGGCACGAAGAGGAGAGGC-- .........(((((..((((((.----.....)))...((((-((..((((---....))))..))))))..))).)))))((((((.(((.....))))))))).............-- ( -32.20) >pf5.2596 4971760 101 + 7074893 GAUUGUUAUCGAGCCGAUCCAUUUUUUGGAUCGGCUUUUUUAUUGCCCGGAGUUCUUCACCGGCAAUAAAAAACCCGCCGCUACCAGCAGGUAUCGGCGGG------------------- (((....)))(((((((((((.....)))))))))))((((((((((.((.(.....).))))))))))))..(((((((.((((....)))).)))))))------------------- ( -52.50) >pfo.2361 4537213 98 + 6438405 GAUUGUUAUCAAGCCGACCCGUUUUUCGGGUCGGCUUUUUUG-UGCCUGCAGUUUUCUGCGGGCAAUAAAAAACCCGCCACGGC--GCAAGCCUCGGCGGG------------------- (((....)))(((((((((((.....)))))))))))(((((-(((((((((....)))))))).))))))..((((((..(((--....)))..))))))------------------- ( -60.30) >consensus GAUUGUUAUCAAGCCGACCCAUU____GGGUCGGCUUUUUUA_UGCCCGCA___UUUCGCCGGCAAUAAAAAACCCGCCGCGACC_GCAGGUAUCGGCGGG___________________ ............(((((((((.....)))))))))(((((((.((((.((((....)))).)))).)))))))(((((((.((((....)))).)))))))................... (-22.60 = -25.60 + 3.00)

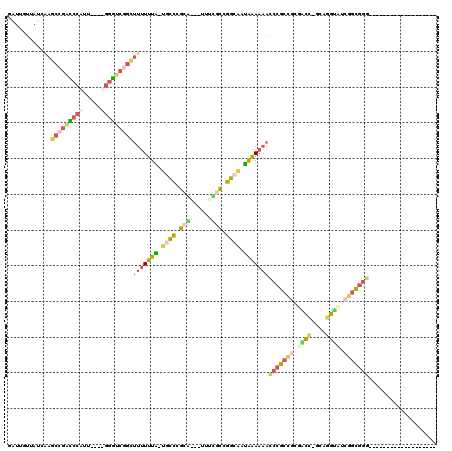
| Location | 3,207,177 – 3,207,289 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 40.17 |
| Mean single sequence MFE | -46.25 |
| Consensus MFE | -15.88 |
| Energy contribution | -21.50 |
| Covariance contribution | 5.62 |
| Combinations/Pair | 1.50 |
| Mean z-score | -4.47 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952255 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>pao.1712 3207177 112 - 6264404 CGGAGAGCGUCGAGGCAGAGGCCCGCGUACCCGCAAGGGACGCGGGUUUUUUCAUGCCCGGCGAAA---CGCAGGCA-UAAAAAAACCGGCCC----CGAGGGACCGGUUUUCAACAGGU ((.....(((((.(((((((((((((((.(((....))))))))))))))....)))))))))...---))......-....(((((((((((----...))).))))))))........ ( -55.20) >pau.1282 4175443 110 + 6537648 --GCCUCUCCUCUUCGUGCCCGGCAGUCUACCAAGGCACCCCGCGGACUUUCCUCCGACGACCUAA---GCCGAUGA-AAAAGUUAAAAAAAU----AUUUGCCAAGCGGGAAAUGUUCG --..........(((((...((((.((((....))))....((((((......)))).))......---))))))))-)...........(((----((((.((....)).))))))).. ( -24.70) >pf5.2596 4971760 101 - 7074893 -------------------CCCGCCGAUACCUGCUGGUAGCGGCGGGUUUUUUAUUGCCGGUGAAGAACUCCGGGCAAUAAAAAAGCCGAUCCAAAAAAUGGAUCGGCUCGAUAACAAUC -------------------(((((((.((((....)))).)))))))((((((((((((.(.((.....))).))))))))))))(((((((((.....)))))))))............ ( -52.60) >pfo.2361 4537213 98 - 6438405 -------------------CCCGCCGAGGCUUGC--GCCGUGGCGGGUUUUUUAUUGCCCGCAGAAAACUGCAGGCA-CAAAAAAGCCGACCCGAAAAACGGGUCGGCUUGAUAACAAUC -------------------(((((((.(((....--))).)))))))........((((.((((....)))).))))-.....(((((((((((.....))))))))))).......... ( -52.50) >consensus ___________________CCCGCCGAUACCCGC_GGACGCGGCGGGUUUUUCAUUGCCGGCGAAA___UCCAGGCA_UAAAAAAACCGACCC____AAUGGGUCGGCUUGAUAACAAUC ...................(((((((.((((....)))).)))))))..(((((.((((.(((......))).)))).)))))(((((((((((.....))))))))))).......... (-15.88 = -21.50 + 5.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:28 2007