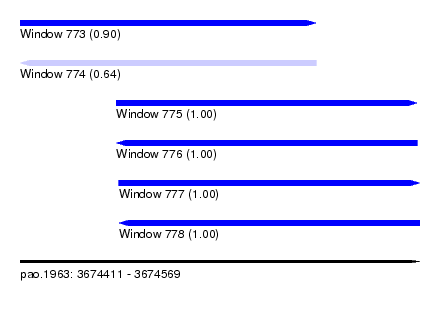
| Sequence ID | pao.1963 |
|---|---|
| Location | 3,674,411 – 3,674,569 |
| Length | 158 |
| Max. P | 0.999922 |

| Location | 3,674,411 – 3,674,528 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -26.91 |
| Energy contribution | -29.03 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1963 3674411 117 + 6264404 GGCCCCUUUGUGUUGGG-GG-AAUCUUGGCGAUGGCAGAUUGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCU (((...(((((((((..-..-....(((((....((.....)).))))).(((((((((((((((.(((((........)))))..))))))))).)))))).-)))))))))..))).. ( -43.10) >pau.1017 4664836 118 - 6537648 GGCCCCUUUGUGUUGGG-GGGAAUCUUGGCGAUGGCAGAUUGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCU (((...(((((((((..-.......(((((....((.....)).))))).(((((((((((((((.(((((........)))))..))))))))).)))))).-)))))))))..))).. ( -43.10) >pf5.919 5351374 119 - 7074893 GGCCCGCUGGUGUUGGGUGGGAAUCUUGGCGAGGGGUUAUUUCAGCCGGGUUGCUGAUAGAUAAAGAUUUGAUUACAGGGGAACUUUUUUACUUCGAAAUGAAAUAAGGCAAAAAGCC-U ..((((((.......))))))..((....(((((..((((((((((......)))))..)))))............(((....))).....)))))....))....((((.....)))-) ( -27.60) >consensus GGCCCCUUUGUGUUGGG_GGGAAUCUUGGCGAUGGCAGAUUGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA_UAACGCGAAAAGCCCU (((...(((((((((..........(((((....((.....)).))))).(((((((((((((((.(((((........)))))..))))))))).))))))..)))))))))..))).. (-26.91 = -29.03 + 2.12)

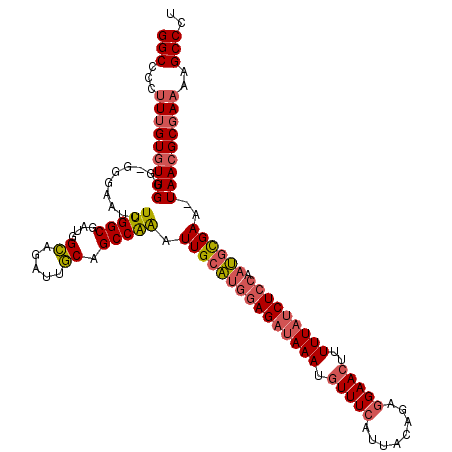
| Location | 3,674,411 – 3,674,528 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -17.68 |
| Energy contribution | -19.47 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1963 3674411 117 - 6264404 AGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCAAUCUGCCAUCGCCAAGAUU-CC-CCCAACACAAAGGGGCC .((..((((.(((...-...((((.(((((((((...(((.........)))...)))))))))))))....((((........)))).))).))))..-))-(((........)))... ( -34.10) >pau.1017 4664836 118 + 6537648 AGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCAAUCUGCCAUCGCCAAGAUUCCC-CCCAACACAAAGGGGCC .((..((((.(((...-...((((.(((((((((...(((.........)))...)))))))))))))....((((........)))).))).))))..))(-(((........)))).. ( -36.50) >pf5.919 5351374 119 + 7074893 A-GGCUUUUUGCCUUAUUUCAUUUCGAAGUAAAAAAGUUCCCCUGUAAUCAAAUCUUUAUCUAUCAGCAACCCGGCUGAAAUAACCCCUCGCCAAGAUUCCCACCCAACACCAGCGGGCC .-(((.....)))(((((((.....))))))).......((((((..................(((((......))))).........((.....))..............))).))).. ( -21.20) >consensus AGGGCUUUUCGCGUUA_UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCAAUCUGCCAUCGCCAAGAUUCCC_CCCAACACAAAGGGGCC ..(((((((.((........((((.(((((((((...(((.........)))...))))))))))))).....(((........)))...)).))))......(((........)))))) (-17.68 = -19.47 + 1.78)

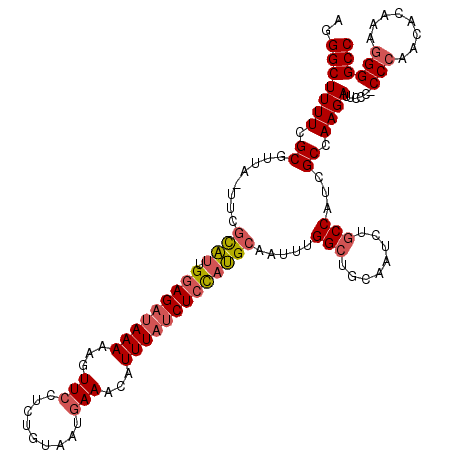
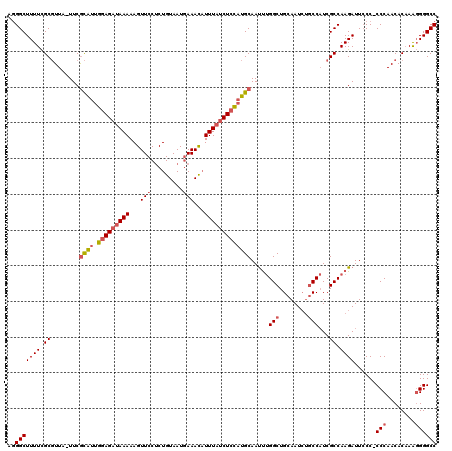
| Location | 3,674,449 – 3,674,568 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.51 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -18.26 |
| Energy contribution | -22.93 |
| Covariance contribution | 4.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1963 3674449 119 + 6264404 UGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAAU .....(((.((((((((((((((((.(((((........)))))..))))))))).))))(((-((((.(((((.((((........)))).))))).)))))))))).)))........ ( -48.10) >pau.1017 4664875 118 - 6537648 UGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAA- .....(((.((((((((((((((((.(((((........)))))..))))))))).))))(((-((((.(((((.((((........)))).))))).)))))))))).))).......- ( -48.10) >pf5.919 5351414 93 - 7074893 UUCAGCCGGGUUGCUGAUAGAUAAAGAUUUGAUUACAGGGGAACUUUUUUACUUCGAAAUGAAAUAAGGCAAAAAGCC-UGUACUA-GAGCCUUU------------------------- .(((((......))))).....((((..((...((((((....((((((..(((...........)))..))))))))-))))...-))..))))------------------------- ( -16.30) >consensus UGCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA_UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAA_ ..........(((((((((((((((.(((((........)))))..))))))))).))))))..((((.(((((.((((........)))).))))).)))).................. (-18.26 = -22.93 + 4.67)

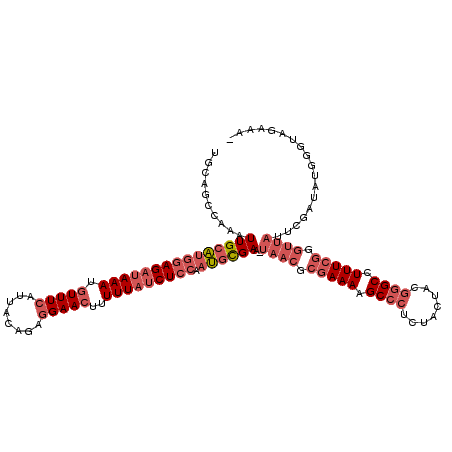
| Location | 3,674,449 – 3,674,568 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.51 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -19.17 |
| Energy contribution | -22.40 |
| Covariance contribution | 3.23 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1963 3674449 119 - 6264404 AUUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCA ........(((....(((((((.((((((((((........)))))))))).))))-)))((((.(((((((((...(((.........)))...)))))))))))))....)))..... ( -44.70) >pau.1017 4664875 118 + 6537648 -UUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCA -.......(((....(((((((.((((((((((........)))))))))).))))-)))((((.(((((((((...(((.........)))...)))))))))))))....)))..... ( -44.70) >pf5.919 5351414 93 + 7074893 -------------------------AAAGGCUC-UAGUACA-GGCUUUUUGCCUUAUUUCAUUUCGAAGUAAAAAAGUUCCCCUGUAAUCAAAUCUUUAUCUAUCAGCAACCCGGCUGAA -------------------------(((((...-...((((-((((((((....((((((.....))))))))))))....))))))......))))).....(((((......))))). ( -20.20) >consensus _UUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA_UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGCA ..................((((.((((((((((........)))))))))).))))....((((.(((((((((...(((.........)))...)))))))))))))............ (-19.17 = -22.40 + 3.23)

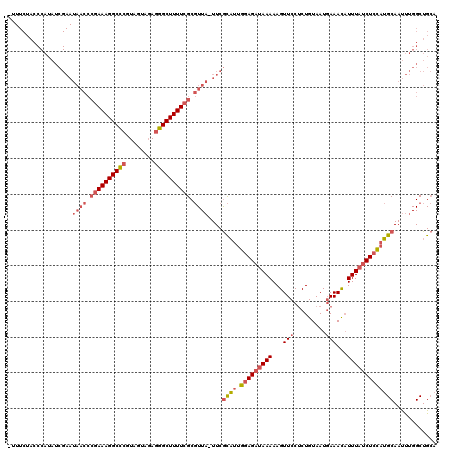
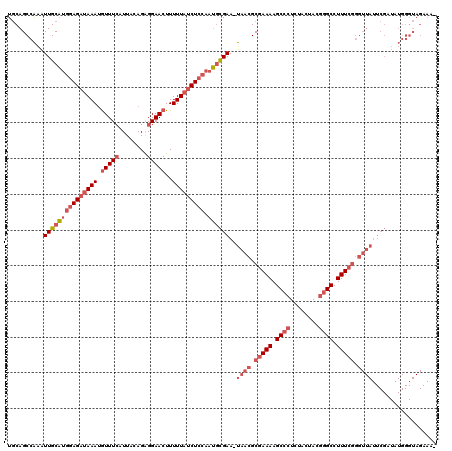
| Location | 3,674,450 – 3,674,569 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -18.26 |
| Energy contribution | -22.93 |
| Covariance contribution | 4.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
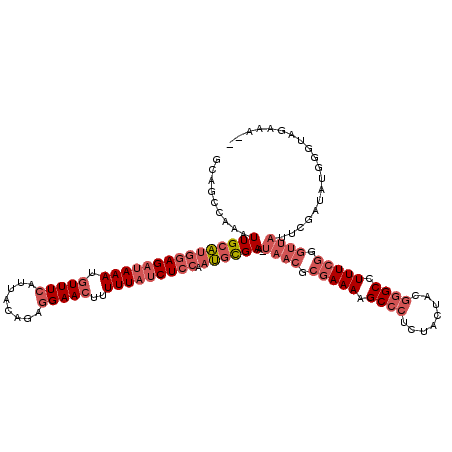
>pao.1963 3674450 119 + 6264404 GCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAAUC ....(((.((((((((((((((((.(((((........)))))..))))))))).))))(((-((((.(((((.((((........)))).))))).)))))))))).)))......... ( -48.10) >pau.1017 4664876 117 - 6537648 GCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA-UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAA-- ....(((.((((((((((((((((.(((((........)))))..))))))))).))))(((-((((.(((((.((((........)))).))))).)))))))))).))).......-- ( -48.10) >pf5.919 5351415 92 - 7074893 UCAGCCGGGUUGCUGAUAGAUAAAGAUUUGAUUACAGGGGAACUUUUUUACUUCGAAAUGAAAUAAGGCAAAAAGCC-UGUACUA-GAGCCUUU-------------------------- (((((......))))).....((((..((...((((((....((((((..(((...........)))..))))))))-))))...-))..))))-------------------------- ( -16.10) >consensus GCAGCCAAAUUGCAUGGAGAUAAAUGUUUCAUUACAGAGGAACUUUUUAUCUCCAAUGCGAA_UAACGCGAAAAGCCCUCUACUACGGGCCUUUCGGGUUAUUCGAUAUGGGUAGAAA__ .........(((((((((((((((.(((((........)))))..))))))))).))))))..((((.(((((.((((........)))).))))).))))................... (-18.26 = -22.93 + 4.67)

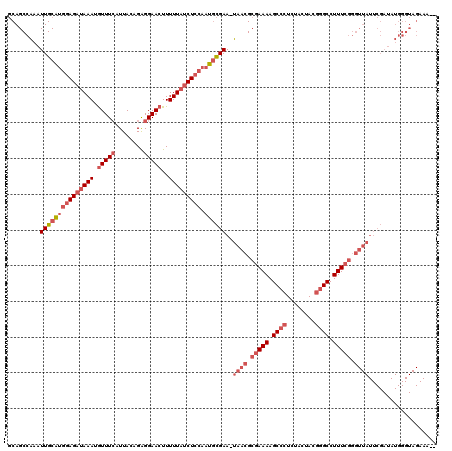
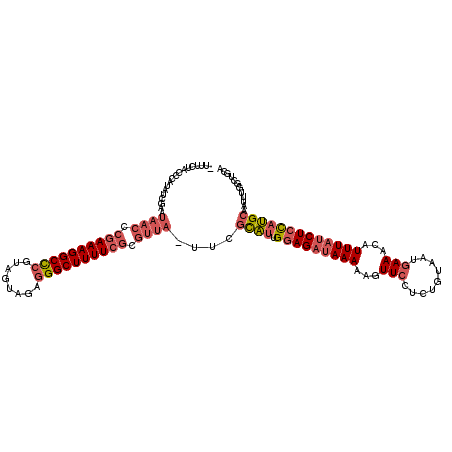
| Location | 3,674,450 – 3,674,569 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -19.17 |
| Energy contribution | -22.40 |
| Covariance contribution | 3.23 |
| Combinations/Pair | 1.13 |
| Mean z-score | -4.56 |
| Structure conservation index | 0.53 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
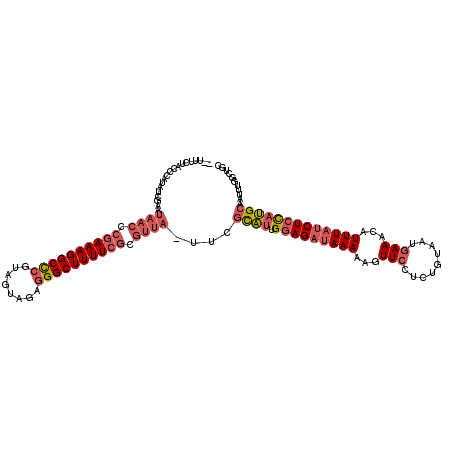
>pao.1963 3674450 119 - 6264404 GAUUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGC .........(((....(((((((.((((((((((........)))))))))).))))-)))((((.(((((((((...(((.........)))...)))))))))))))....))).... ( -44.70) >pau.1017 4664876 117 + 6537648 --UUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA-UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGC --.......(((....(((((((.((((((((((........)))))))))).))))-)))((((.(((((((((...(((.........)))...)))))))))))))....))).... ( -44.70) >pf5.919 5351415 92 + 7074893 --------------------------AAAGGCUC-UAGUACA-GGCUUUUUGCCUUAUUUCAUUUCGAAGUAAAAAAGUUCCCCUGUAAUCAAAUCUUUAUCUAUCAGCAACCCGGCUGA --------------------------(((((...-...((((-((((((((....((((((.....))))))))))))....))))))......))))).....(((((......))))) ( -19.40) >consensus __UUUCUACCCAUAUCGAAUAACCCGAAAGGCCCGUAGUAGAGGGCUUUUCGCGUUA_UUCGCAUUGGAGAUAAAAAGUUCCUCUGUAAUGAAACAUUUAUCUCCAUGCAAUUUGGCUGC ...................((((.((((((((((........)))))))))).))))....((((.(((((((((...(((.........)))...)))))))))))))........... (-19.17 = -22.40 + 3.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:19 2007