
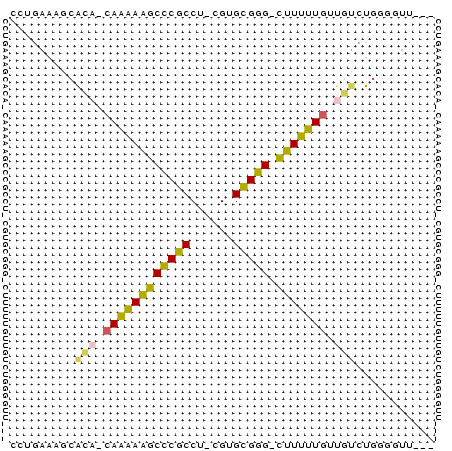
| Sequence ID | pao.3062 |
|---|---|
| Location | 5,798,634 – 5,798,692 |
| Length | 58 |
| Max. P | 0.999300 |

| Location | 5,798,634 – 5,798,692 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -16.28 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.53 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3062 5798634 58 + 6264404 CAUGAAUACGAC-GAAAAGGCUCACCUCCGGGUGGGCCUUUUUGCUUUCCGCCGUUCGC ...((((.((.(-(((((((((((((....)))))))))))))).....))..)))).. ( -23.90) >pss.2982 5816479 53 + 6093698 CAUGAAAGCACA-CAAAAAGCCCGCCU-CGUGCGGG-CUUUUUGCUGUCUGGGGUU--- ......((.(((-((((((((((((..-...)))))-))))))).)))))......--- ( -22.70) >ppk.165 5837795 54 - 6181863 CCCGAAGCGACAACAAGAAACCCGCCU-AGUGCGGG-UUUUUUGUUGUCUGUGAUU--- ......(((((((((((((((((((..-...)))))-)))))))))))).))....--- ( -27.90) >pf5.219 6665929 53 - 7074893 CGUGAAAGCAAC-CAGAAAACCCGCCU-AGUGCGGG-UUUUUUGUUGCCUGAGAUU--- .......(((((-.(((((((((((..-...)))))-)))))))))))........--- ( -22.30) >psp.2878 5605680 50 + 5928787 CCUGAAAGCACA-CAAGAAGCCCGCCU-CGUGCGGG-CUUUUUGUUGUCUGGG------ ((((...(((.(-((((((((((((..-...)))))-))))))))))).))))------ ( -23.80) >pst.3180 6074648 52 + 6397126 CCUGAAA-CACA-CAAGAAGCCCGCCU-CGUGCGGG-CUUUUUGUUGUCUGGAGUU--- ((....(-((.(-((((((((((((..-...)))))-)))))))))))..))....--- ( -23.90) >consensus CCUGAAAGCACA_CAAAAAGCCCGCCU_CGUGCGGG_CUUUUUGUUGUCUGGGGUU___ .........(((.((((((((((((......))))).))))))).)))........... (-16.28 = -15.40 + -0.88)


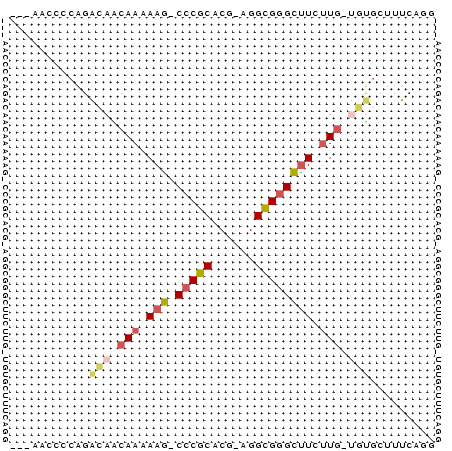
| Location | 5,798,634 – 5,798,692 |
|---|---|
| Length | 58 |
| Sequences | 6 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3062 5798634 58 - 6264404 GCGAACGGCGGAAAGCAAAAAGGCCCACCCGGAGGUGAGCCUUUUC-GUCGUAUUCAUG ..(((((((((((.......((((.((((....)))).))))))))-)))))..))... ( -21.21) >pss.2982 5816479 53 - 6093698 ---AACCCCAGACAGCAAAAAG-CCCGCACG-AGGCGGGCUUUUUG-UGUGCUUUCAUG ---......((...((((((((-(((((...-..))))))))))))-)...))...... ( -23.40) >ppk.165 5837795 54 + 6181863 ---AAUCACAGACAACAAAAAA-CCCGCACU-AGGCGGGUUUCUUGUUGUCGCUUCGGG ---.......((((((((.(((-(((((...-..)))))))).)))))))).(....). ( -25.90) >pf5.219 6665929 53 + 7074893 ---AAUCUCAGGCAACAAAAAA-CCCGCACU-AGGCGGGUUUUCUG-GUUGCUUUCACG ---......(((((((..((((-(((((...-..)))))))))...-)))))))..... ( -22.40) >psp.2878 5605680 50 - 5928787 ------CCCAGACAACAAAAAG-CCCGCACG-AGGCGGGCUUCUUG-UGUGCUUUCAGG ------...((...((((.(((-(((((...-..)))))))).)))-)...))...... ( -19.60) >pst.3180 6074648 52 - 6397126 ---AACUCCAGACAACAAAAAG-CCCGCACG-AGGCGGGCUUCUUG-UGUG-UUUCAGG ---..((..(((((((((.(((-(((((...-..)))))))).)))-).))-)))..)) ( -21.70) >consensus ___AACCCCAGACAACAAAAAG_CCCGCACG_AGGCGGGCUUCUUG_UGUGCUUUCAGG ...........(((.(((.(((.(((((......)))))))).))).)))......... (-11.10 = -11.38 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:25 2007