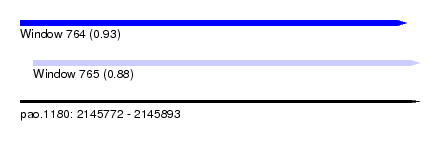
| Sequence ID | pao.1180 |
|---|---|
| Location | 2,145,772 – 2,145,893 |
| Length | 121 |
| Max. P | 0.926939 |

| Location | 2,145,772 – 2,145,889 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.17 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -18.50 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
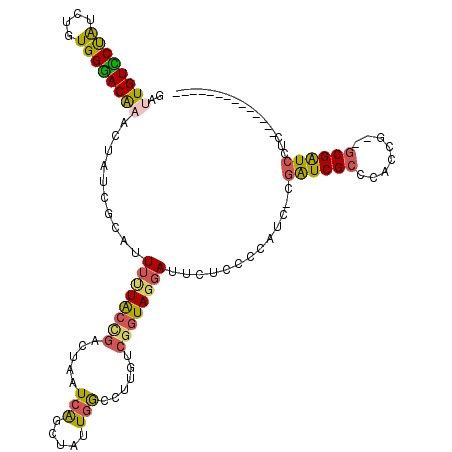
>pao.1180 2145772 117 + 6264404 GAUUGUCCUAUGUGUGGGACGAAUAGUCAUGUUUCUACCGACUAAUCGUCGAUUGGCCUUGCCGGUAGGAUGCAAACCACU-CGAUCGCCACACU--GCGAUCCCCCACUUUCAGGAGCG .....((((..(.(((((((.....))).(((((((((((((.....)))....(((...)))))))))).)))..)))).-)((((((......--))))))..........))))... ( -38.30) >pau.1845 3049206 117 - 6537648 GAUUGUCCUAUGUGUGGGACGAAUAGUCAUGUUUCUACCGACUAAUCGUCGAUUGGCCUUGCCGGUAGGAUGCAAACCACU-CGAUCGCCACACU--GCGAUCCCCCACUUUCAGGAGCG .....((((..(.(((((((.....))).(((((((((((((.....)))....(((...)))))))))).)))..)))).-)((((((......--))))))..........))))... ( -38.30) >pf5.1863 3491632 103 - 7074893 GAUUGUCCUGCUGAUGGGACGAACUAUCGCAUUUAUACCGGCUAAUCAGCUAUUGGUUUCAUCUGUAGGAUUGCCCACAUC-CGAUCGCCCAGUG--GCGGUCCUC-------------- ....(((((((.((((..((((....))...........((((....))))....))..)))).)))))))..........-.(((((((....)--))))))...-------------- ( -33.10) >pfo.1521 2982846 102 + 6438405 GACUGUUCUAUCAGUGGGACAAACUAUCGCAUUGUUGCCGUCUAAUCAGCUAUUGGUUUCGUCGGUAGGAUUGUCUCCAUU-CCGUCG-CCACCG--GCGAUUCAU-------------- .((((......))))(((((((.((((((...((..((((.............))))..)).))))))..))))).))...-..((((-((...)--)))))....-------------- ( -29.12) >ppk.1654 3267404 105 + 6181863 -ACUGUUCCAUCUGUGGGACAAACUAACGCAUUUUUGCUUACUUAUCAGCUAUUGAUCAUCUCGGUAGGAUUCUCCCCAUCGUGAUCGCCCACAGCUGCGGUCUUC-------------- -(((((.....(((((((.(........(((....)))................(((((...((((.((......)).)))))))))))))))))..)))))....-------------- ( -28.30) >pel.1675 2806684 105 - 5888780 -AUUGUUCCAUCAGUGGAACAAACUAACGCAUUUUUGCUGGCUUAUCAGCCAUUCGCUGUAUCGAUAGGAUUCUCCCCAUCGUGAUCGCCCACCGCCGCGGUCUUC-------------- -.((((((((....))))))))......(((....)))(((((....)))))..........((((.((......)).)))).((((((........))))))...-------------- ( -30.10) >consensus GAUUGUCCUAUCUGUGGGACAAACUAUCGCAUUUUUACCGACUAAUCAGCUAUUGGCCUUGUCGGUAGGAUUCUCCCCAUC_CGAUCGCCCACCG__GCGAUCCUC______________ ..((((((((....))))))))..........((((((((.....(((.....)))......)))))))).............((((((........))))))................. (-18.50 = -17.83 + -0.66)

| Location | 2,145,776 – 2,145,893 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.91 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -17.50 |
| Energy contribution | -16.75 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
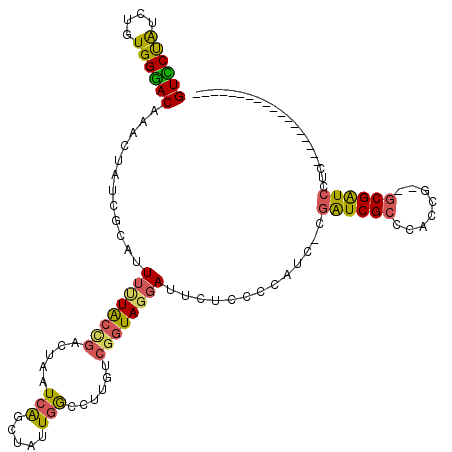
>pao.1180 2145776 117 + 6264404 GUCCUAUGUGUGGGACGAAUAGUCAUGUUUCUACCGACUAAUCGUCGAUUGGCCUUGCCGGUAGGAUGCAAACCACU-CGAUCGCCACACU--GCGAUCCCCCACUUUCAGGAGCGAGAC .((((..(.(((((((.....))).(((((((((((((.....)))....(((...)))))))))).)))..)))).-)((((((......--))))))..........))))....... ( -38.30) >pau.1845 3049210 115 - 6537648 GUCCUAUGUGUGGGACGAAUAGUCAUGUUUCUACCGACUAAUCGUCGAUUGGCCUUGCCGGUAGGAUGCAAACCACU-CGAUCGCCACACU--GCGAUCCCCCACUUUCAGGAGCGAG-- .((((..(.(((((((.....))).(((((((((((((.....)))....(((...)))))))))).)))..)))).-)((((((......--))))))..........)))).....-- ( -38.30) >pf5.1863 3491636 99 - 7074893 GUCCUGCUGAUGGGACGAACUAUCGCAUUUAUACCGGCUAAUCAGCUAUUGGUUUCAUCUGUAGGAUUGCCCACAUC-CGAUCGCCCAGUG--GCGGUCCUC------------------ (((((((.((((..((((....))...........((((....))))....))..)))).)))))))..........-.(((((((....)--))))))...------------------ ( -32.90) >pfo.1521 2982850 98 + 6438405 GUUCUAUCAGUGGGACAAACUAUCGCAUUGUUGCCGUCUAAUCAGCUAUUGGUUUCGUCGGUAGGAUUGUCUCCAUU-CCGUCG-CCACCG--GCGAUUCAU------------------ ........((((((((((.((((((...((..((((.............))))..)).))))))..))))).)))))-..((((-((...)--)))))....------------------ ( -28.32) >ppk.1654 3267407 102 + 6181863 GUUCCAUCUGUGGGACAAACUAACGCAUUUUUGCUUACUUAUCAGCUAUUGAUCAUCUCGGUAGGAUUCUCCCCAUCGUGAUCGCCCACAGCUGCGGUCUUC------------------ ..((((.(((((((.(........(((....)))................(((((...((((.((......)).))))))))))))))))).)).)).....------------------ ( -26.50) >pel.1675 2806687 102 - 5888780 GUUCCAUCAGUGGAACAAACUAACGCAUUUUUGCUGGCUUAUCAGCCAUUCGCUGUAUCGAUAGGAUUCUCCCCAUCGUGAUCGCCCACCGCCGCGGUCUUC------------------ ((((((....))))))........(((....)))(((((....)))))..........((((.((......)).)))).((((((........))))))...------------------ ( -28.20) >consensus GUCCUAUCUGUGGGACAAACUAUCGCAUUUUUACCGACUAAUCAGCUAUUGGCCUUGUCGGUAGGAUUCUCCCCAUC_CGAUCGCCCACCG__GCGAUCCUC__________________ ((((((....))))))............((((((((.....(((.....)))......)))))))).............((((((........))))))..................... (-17.50 = -16.75 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:05 2007