| Sequence ID | pao.954 |
|---|---|
| Location | 1,711,191 – 1,711,298 |
| Length | 107 |
| Max. P | 0.825121 |

| Location | 1,711,191 – 1,711,298 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.95 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
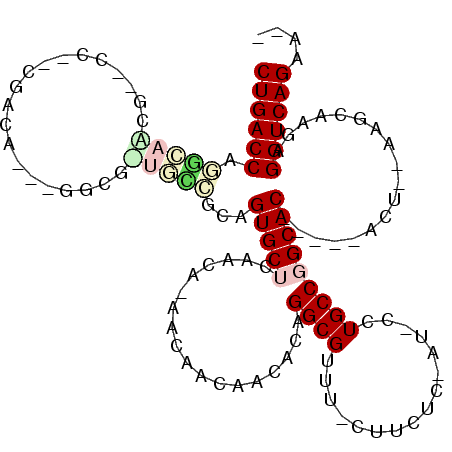
>pao.954 1711191 107 - 6264404 CUGACCAGGCACCG------CGACA--UGGCGGUGCGGAAGUGCUCAUCAGAACAACAACA-AGGCGUGUUCCCCUC-ACACCUGCCGGCACCGAUAAC---AAGAUCGAGGUCAGAACC ((((((..((((((------(....--..)))))))....((((((....)).........-.(((((((.......-))))..))).))))((((...---...)))).)))))).... ( -37.80) >pfo.2155 4165489 108 - 6438405 CUGACC-GGUAUCG--CCGAAGGGACGGGGUGAUAUCGCAGUGCUCAACA-AUAAAAAACACAGGCGUUUACUUAUCGAU-UCUGCCGGCAC-----ACUCACACCCUGAGGUCAGAA-- ((((((-......(--(((..((((((((((((...(((.(((.......-........)))..))).)))))..))).)-)))..))))..-----..(((.....)))))))))..-- ( -30.96) >pel.1387 3399507 98 + 5888780 CUGACCAGGUGACGUCCCUCCGUCA-----------CGCAGUGCGCAACACAACAACAAGACAGGCGUUU-CUUCAC-AU-UCUGCCGGCAC-----ACUC-AAGCAAGAGGUCAGAU-- ((((((..((((((......)))))-----------)((((((.((.......(.....)...((((...-......-..-..)))).)).)-----))).-..))....))))))..-- ( -29.12) >pf5.2400 4575817 104 - 7074893 CUGACC-AGCAGCG-CCCGCCGCGA--GGGCCCUGCUGCAGUGCUCAAGA-AUAAAAAACACAGGCGUUUGCUUCGC-AU-CCUGCCGGCAC-----ACU--CACCCUGAGGUCAGAA-- ((((((-(((((.(-(((.......--)))).)))))...(((((.....-............((((..(((...))-).-.).))))))))-----...--........))))))..-- ( -37.52) >pau.2074 2605336 105 + 6537648 CUGACCAAGCACCG------CGACA--UGGCGGUGCGGAAGUGCUCAUCAGAACAACAACA-AGGCGUGUUCCCCUC-ACACCUGCCGGCACCGAUAAC---AAGAUCGAGGUCAGAA-- ((((((..((((((------(....--..)))))))....((((((....)).........-.(((((((.......-))))..))).))))((((...---...)))).))))))..-- ( -38.00) >ppk.1491 3288861 96 + 6181863 CUGACCCGAUGACAGCCU--CGUCA----------UCGCUGUGCGUAACACAAGAACAAGACAGGCGUUU-CUUCAC-AU-UCUGCCGGCAC-----ACUU-GAGCAAGAGGUCAGA--- (((((((((((((.....--.))))----------))((((((.....))).....((((...((((...-......-..-..)))).....-----.)))-))))..).)))))).--- ( -28.82) >consensus CUGACCAGGCAACG__CC__CGACA___GGCG_UGCCGCAGUGCUCAACA_AACAACAACACAGGCGUUU_CUUCUC_AU_CCUGCCGGCAC_____ACU__AAGCAAGAGGUCAGAA__ ((((((.(((((....................)))))...(((((..................((((................)))))))))..................)))))).... (-14.02 = -14.97 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:17:00 2007