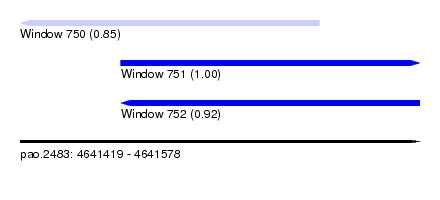
| Sequence ID | pao.2483 |
|---|---|
| Location | 4,641,419 – 4,641,578 |
| Length | 159 |
| Max. P | 0.998086 |

| Location | 4,641,419 – 4,641,538 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -62.37 |
| Consensus MFE | -43.42 |
| Energy contribution | -41.43 |
| Covariance contribution | -1.98 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2483 4641419 119 - 6264404 GUCCGGC-GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUGGCGGCCCGGCGCUUGUCGAGGGUCGCCGAUGGGCGCUUAA (((((((-((.....(((.((.((..((((.((((((((((((..((((((((((((...)))))))).)))).)))).)).)))))))))).)).)).))))))))...))))...... ( -63.90) >pau.477 5651212 119 + 6537648 GUCCGGC-GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUGGCGGCCCGGCGCUUGUCGAGGGUCGCCGAUGGGCGCUUAA (((((((-((.....(((.((.((..((((.((((((((((((..((((((((((((...)))))))).)))).)))).)).)))))))))).)).)).))))))))...))))...... ( -63.90) >pf5.1312 2424931 120 - 7074893 CUGCGCCUGAGACGUUGCAUGUCAGGCUGCACGGGGCCUUGGCGAGACCCUGAGACGUUGCGUCUCAGCGCCCCCACGGCGCCGCCCGGUGCUUGUCGGGAGCCGCGGAUCGGCGCUUAA ..(((((.((..(((.((.(.((.(((.((((.((((...(((......((((((((...)))))))).(((.....)))))))))).))))..))))).))).)))..))))))).... ( -59.30) >consensus GUCCGGC_GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUGGCGGCCCGGCGCUUGUCGAGGGUCGCCGAUGGGCGCUUAA ((((.((...((((.....)))).((.(((.(((....)))))).((((((((((((...)))))))).))))))..)).).)))..((((((..(((........)))..))))))... (-43.42 = -41.43 + -1.98)


| Location | 4,641,459 – 4,641,578 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.82 |
| Mean single sequence MFE | -61.70 |
| Consensus MFE | -40.47 |
| Energy contribution | -40.70 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2483 4641459 119 + 6264404 CACAGCCGACGACUGAGACGGAGCGUCUCAGCGUCUCGCCGGGAACCCGUGCGCGAUGACAUGGGUUGUCUC-GCCGGACGUCGCCCCGGCGGUCGCCGGCCUUCCCAUGCCGGCGCUCU ....(((((((.((((((((...))))))))))))..((((((....(((.(((((.((((.....))))))-).)).)))....)))))))))(((((((........))))))).... ( -63.40) >pau.477 5651252 119 - 6537648 CACAGCCGACGACUGAGACGGAGCGUCUCAGCGUCUCGCCGGGAACCCGUGCGCGAUGACAUGGGUUGUCUC-GCCGGACGUCGCCCCGGCGGUCGCCGGCCUUCCCAUGCCGGCGCUCU ....(((((((.((((((((...))))))))))))..((((((....(((.(((((.((((.....))))))-).)).)))....)))))))))(((((((........))))))).... ( -63.40) >pf5.1312 2424971 119 + 7074893 GCCGUGGGGGCGCUGAGACGCAACGUCUCAGGGUCUCGCCAAGGCCCCGUGCAGCCUGACAUGCAACGUCUCAGGCGCAGGGCUUCCCGACACCUGCAGGCGAGUGCA-ACAGCCGCUCU ((((..(((((.((((((((...)))))))).)))(((...((((((..(((.((((((.(((...))).)))))))))))))))..)))..))..).)))((((((.-...).))))). ( -58.30) >consensus CACAGCCGACGACUGAGACGGAGCGUCUCAGCGUCUCGCCGGGAACCCGUGCGCGAUGACAUGGGUUGUCUC_GCCGGACGUCGCCCCGGCGGUCGCCGGCCUUCCCAUGCCGGCGCUCU ..(((((((((.((((((((...))))))))))))..(((((....))).))...........))))).....(((((........(((((....)))))..........)))))..... (-40.47 = -40.70 + 0.23)

| Location | 4,641,459 – 4,641,578 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.82 |
| Mean single sequence MFE | -61.10 |
| Consensus MFE | -43.47 |
| Energy contribution | -42.27 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2483 4641459 119 - 6264404 AGAGCGCCGGCAUGGGAAGGCCGGCGACCGCCGGGGCGACGUCCGGC-GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUG ..(((((((((........))))))...((((((((...(((.((.(-((((((.....)))).))))).)))...)))))))).((((((((((((...)))))))).)))).)))... ( -64.30) >pau.477 5651252 119 + 6537648 AGAGCGCCGGCAUGGGAAGGCCGGCGACCGCCGGGGCGACGUCCGGC-GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUG ..(((((((((........))))))...((((((((...(((.((.(-((((((.....)))).))))).)))...)))))))).((((((((((((...)))))))).)))).)))... ( -64.30) >pf5.1312 2424971 119 - 7074893 AGAGCGGCUGU-UGCACUCGCCUGCAGGUGUCGGGAAGCCCUGCGCCUGAGACGUUGCAUGUCAGGCUGCACGGGGCCUUGGCGAGACCCUGAGACGUUGCGUCUCAGCGCCCCCACGGC ...((((((..-((((..((.((.(((((((.(((...))).))))))))).)).)))).....)))))).(((((....((((.....((((((((...))))))))))))))).)).. ( -54.70) >consensus AGAGCGCCGGCAUGGGAAGGCCGGCGACCGCCGGGGCGACGUCCGGC_GAGACAACCCAUGUCAUCGCGCACGGGUUCCCGGCGAGACGCUGAGACGCUCCGUCUCAGUCGUCGGCUGUG ...((((.((((((((....(((((....)))))(....)...............))))))))...)))).(((....)))....((((((((((((...)))))))).))))....... (-43.47 = -42.27 + -1.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:51 2007