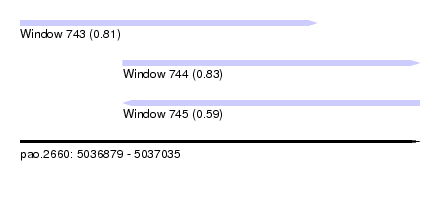
| Sequence ID | pao.2660 |
|---|---|
| Location | 5,036,879 – 5,037,035 |
| Length | 156 |
| Max. P | 0.826520 |

| Location | 5,036,879 – 5,036,995 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.810026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
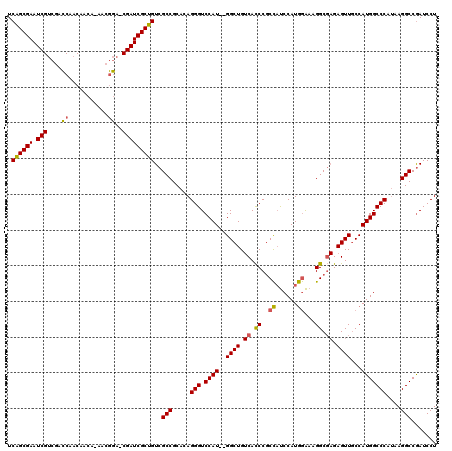
>pao.2660 5036879 116 + 6264404 UUUUCGUUGACAACAAUUUCAGCAUGACUUAAUUUCGGCAUCAGCGAAUCGUCGACCAACAACA-AACGGA-CGAUCGCUGUCGCCGCACAGGGUCCAU--GGCUGUCACCCGCCAUCCA .....(((((........)))))..(((((.....((((..((((((.(((((...........-....))-)))))))))..))))....))))).((--(((........)))))... ( -37.06) >pau.2746 5200462 116 + 6537648 UUUUCGUUGACAACAAUUUCAGCAUGACUUAAUUUCGGCAUCAGCGAAUCGUCGACCAACAACA-AACGGA-CGAUCGCUGUCGCCGCACAGGGUCCAU--GGCUGUCACCCGCCAUCCA .....(((((........)))))..(((((.....((((..((((((.(((((...........-....))-)))))))))..))))....))))).((--(((........)))))... ( -37.06) >pf5.526 6049424 119 - 7074893 UUUUCGUUGACCACGAUUUCAGCAUGACUUAAUUUCGGCAUCGGCGAAUCGACUAUCAACAACACAACAAAGCGAUCGCCGUCGCCGCACAGGGUCCAUGGGGCUG-CAUCCACUCCCCA .....(((((........)))))..(((((.....((((..((((((.(((.((................)))))))))))..))))....)))))..(((((.((-....))..))))) ( -34.89) >consensus UUUUCGUUGACAACAAUUUCAGCAUGACUUAAUUUCGGCAUCAGCGAAUCGUCGACCAACAACA_AACGGA_CGAUCGCUGUCGCCGCACAGGGUCCAU__GGCUGUCACCCGCCAUCCA .....(((((........)))))..(((((.....((((..((((((.(((....((...........))..)))))))))..))))....))))).....(((........)))..... (-28.51 = -28.07 + -0.44)

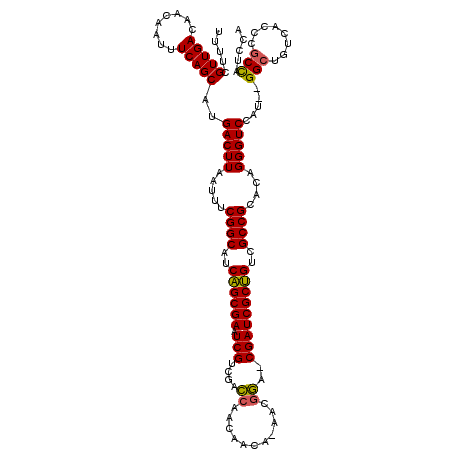
| Location | 5,036,919 – 5,037,035 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -31.13 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
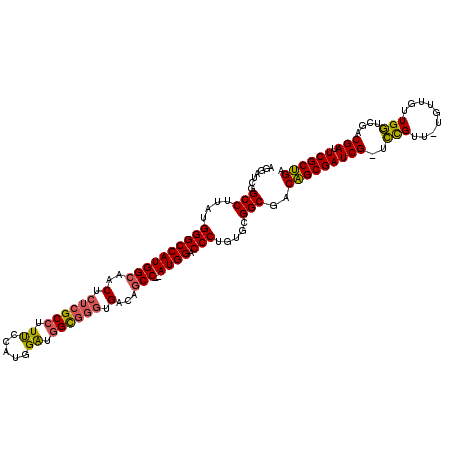
>pao.2660 5036919 116 + 6264404 UCAGCGAAUCGUCGACCAACAACA-AACGGA-CGAUCGCUGUCGCCGCACAGGGUCCAU--GGCUGUCACCCGCCAUCCAUGGAAAGGCGAGAGUUGCCAUGGCCCAUAAGGCCGAUCCU .((((((.(((((...........-....))-)))))))))(((((.....(((.((((--(((...(.(.((((.((....))..)))).).)..))))))))))....)).))).... ( -46.16) >pau.2746 5200502 116 + 6537648 UCAGCGAAUCGUCGACCAACAACA-AACGGA-CGAUCGCUGUCGCCGCACAGGGUCCAU--GGCUGUCACCCGCCAUCCAUGGAAAGGCGAGAGUUGCCAUGGCCCAUAAGGCCGAUCCU .((((((.(((((...........-....))-)))))))))(((((.....(((.((((--(((...(.(.((((.((....))..)))).).)..))))))))))....)).))).... ( -46.16) >pf5.526 6049464 117 - 7074893 UCGGCGAAUCGACUAUCAACAACACAACAAAGCGAUCGCCGUCGCCGCACAGGGUCCAUGGGGCUG-CAUCCACUCCCCAUAAGAAGGC--GAGUUUCCAUGGCCCACAAGGCCAAUCCU .((((((.(((.((................)))))))))))..(((.....(((.((((((((...-...))((((.((.......)).--))))..)))))))))....)))....... ( -37.89) >consensus UCAGCGAAUCGUCGACCAACAACA_AACGGA_CGAUCGCUGUCGCCGCACAGGGUCCAU__GGCUGUCACCCGCCAUCCAUGGAAAGGCGAGAGUUGCCAUGGCCCAUAAGGCCGAUCCU .((((((.(((....((...........))..)))))))))..(((.....(((.((((..((((.((.((..((......))...)).)).))))...)))))))....)))....... (-31.13 = -30.67 + -0.47)

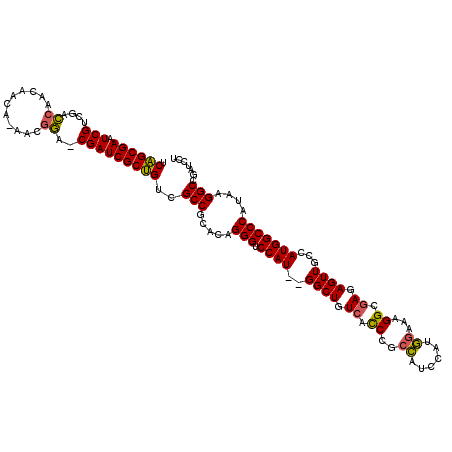
| Location | 5,036,919 – 5,037,035 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -49.87 |
| Consensus MFE | -36.23 |
| Energy contribution | -36.13 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2660 5036919 116 - 6264404 AGGAUCGGCCUUAUGGGCCAUGGCAACUCUCGCCUUUCCAUGGAUGGCGGGUGACAGCC--AUGGACCCUGUGCGGCGACAGCGAUCG-UCCGUU-UGUUGUUGGUCGACGAUUCGCUGA .......(((.(((((((((((((..(.((((((..((....)).)))))).)...)))--)))).))).))).)))..(((((((((-((..((-.......))..))))).)))))). ( -52.50) >pau.2746 5200502 116 - 6537648 AGGAUCGGCCUUAUGGGCCAUGGCAACUCUCGCCUUUCCAUGGAUGGCGGGUGACAGCC--AUGGACCCUGUGCGGCGACAGCGAUCG-UCCGUU-UGUUGUUGGUCGACGAUUCGCUGA .......(((.(((((((((((((..(.((((((..((....)).)))))).)...)))--)))).))).))).)))..(((((((((-((..((-.......))..))))).)))))). ( -52.50) >pf5.526 6049464 117 + 7074893 AGGAUUGGCCUUGUGGGCCAUGGAAACUC--GCCUUCUUAUGGGGAGUGGAUG-CAGCCCCAUGGACCCUGUGCGGCGACGGCGAUCGCUUUGUUGUGUUGUUGAUAGUCGAUUCGCCGA .......(((....(((((((((..((((--.((.......)).))))((...-...)))))))).))).....)))..(((((((((..((((((......)))))).))).)))))). ( -44.60) >consensus AGGAUCGGCCUUAUGGGCCAUGGCAACUCUCGCCUUUCCAUGGAUGGCGGGUGACAGCC__AUGGACCCUGUGCGGCGACAGCGAUCG_UCCGUU_UGUUGUUGGUCGACGAUUCGCUGA .......(((....((((((((((..(.((((((.((.....)).)))))).)...)))..)))).))).....)))..(((((((((..(((.........)))....))).)))))). (-36.23 = -36.13 + -0.10)

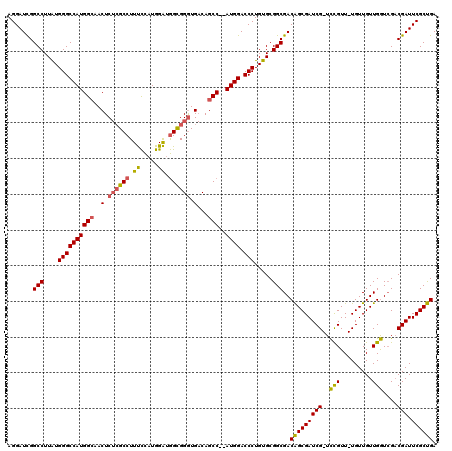
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:43 2007