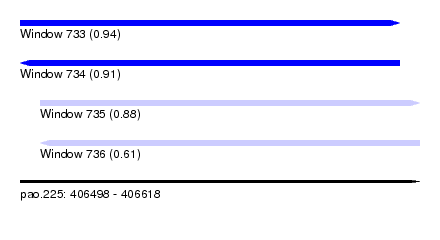
| Sequence ID | pao.225 |
|---|---|
| Location | 406,498 – 406,618 |
| Length | 120 |
| Max. P | 0.935718 |

| Location | 406,498 – 406,612 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -22.23 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
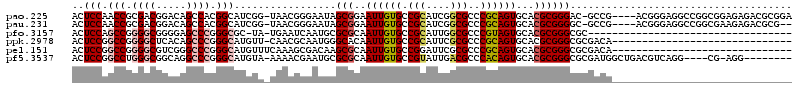
>pao.225 406498 114 + 6264404 CUAACAACUCCAACCGCGACGGACAGCCACGGCAUCGG-UAACGGGAAUAGCGGAAUUGUGCCGCAUCGGCGCCCGCAGUGCACGCGGGAC-GCCG----ACGGGAGGCCGGCGGAGAGA .......(((...((((..(((.(..((.(((((.(((-(..((.......))..)))))))))(.(((((((((((.......))))).)-))))----).)))..)))))))).))). ( -49.60) >pau.231 421252 112 + 6537648 --AACAACUCCAACCGCGACGGACAGCCACGGCAUCGG-UAACGGGAAUAGCGGAAUUGUGCCGCAUCGGCGCCCGCAGUGCACGCGGGGC-GCCG----ACGGGAGGCCGGCGAAGAGA --.....(((....(((..(((.(..((.(((((.(((-(..((.......))..)))))))))(.(((((((((((.......)).))))-))))----).)))..)))))))..))). ( -46.00) >pfo.3157 429187 85 - 6438405 -----AACUCCAGCCGGGGCGGGGAGCCCGGGCGC-UA-UGAAUCAAUGCGCGCAAUUGUGCCGCAUUGGCGCCCGUAGUGCACGCGGGCGC---------------------------- -----.......(((.((((.....)))).)))..-..-.....((((((((((....))).)))))))((((((((.......))))))))---------------------------- ( -44.80) >ppk.2978 336200 89 - 6181863 ------ACUCCGGCCGGGGCUCACAGCCCGGGCAUGUU-CAACGCAAUGGGCACAAUUGUGCCGCAUUCGCGCCCGCAGUGCACGCGGGCGCGACA------------------------ ------......(((.((((.....)))).))).....-.......((((((((....))))).)))((((((((((.......))))))))))..------------------------ ( -47.90) >pel.151 303916 90 + 5888780 ------ACUCCGGCCGGGGCGUCGGGCCCGGGCAUGUUUCAAAGCGACAAGCGCAAUUGUGCCGGAUUCGCGCCCGCAGUGCACGCGGGCGCGACA------------------------ ------..(((((((.((((.....)))).))).................((((....)))))))).((((((((((.......))))))))))..------------------------ ( -47.80) >pf5.3537 400626 106 - 7074893 ------ACUCCGGCCUGGGCGGCAGGCCCGGGCAUGUA-AAAACGAAUGCGCGCAAUUGUGCCGUAUUGACGCCCACAGUGCACGCGGGCGCGAUGGCUGACGUCAGG----CG-AGG-- ------.(((((((((((((.....)))))))).....-....((..((((.(((.(((((.(((....)))..)))))))).))))..))))..(.(((....))).----))-)).-- ( -44.50) >consensus ______ACUCCAGCCGGGGCGGACAGCCCGGGCAUGUA_UAAAGCAAUGAGCGCAAUUGUGCCGCAUUGGCGCCCGCAGUGCACGCGGGCGCGACG________________________ ........(((.(((.((((.....)))).))).................(((..((((((.(((....)))..))))))...))))))............................... (-22.23 = -23.07 + 0.84)

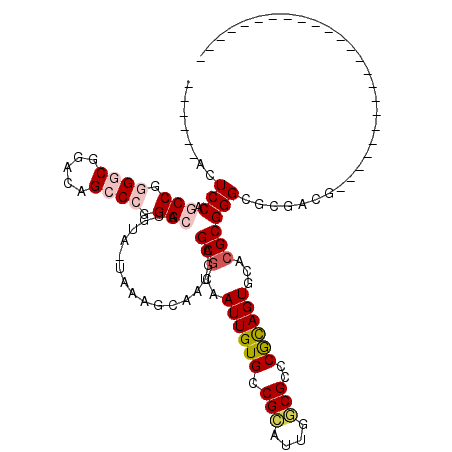
| Location | 406,498 – 406,612 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -45.87 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.225 406498 114 - 6264404 UCUCUCCGCCGGCCUCCCGU----CGGC-GUCCCGCGUGCACUGCGGGCGCCGAUGCGGCACAAUUCCGCUAUUCCCGUUA-CCGAUGCCGUGGCUGUCCGUCGCGGUUGGAGUUGUUAG .......((((......(((----((((-(.((((((.....))))))))))))))))))((((((((........((...-.))..((((((((.....)))))))).))))))))... ( -52.30) >pau.231 421252 112 - 6537648 UCUCUUCGCCGGCCUCCCGU----CGGC-GCCCCGCGUGCACUGCGGGCGCCGAUGCGGCACAAUUCCGCUAUUCCCGUUA-CCGAUGCCGUGGCUGUCCGUCGCGGUUGGAGUUGUU-- .......((((......(((----((((-((((.(((.....))))))))))))))))))((((((((........((...-.))..((((((((.....)))))))).)))))))).-- ( -52.30) >pfo.3157 429187 85 + 6438405 ----------------------------GCGCCCGCGUGCACUACGGGCGCCAAUGCGGCACAAUUGCGCGCAUUGAUUCA-UA-GCGCCCGGGCUCCCCGCCCCGGCUGGAGUU----- ----------------------------(((((((.........)))))))((((((((((....))).)))))))((((.-..-..(((.((((.....)))).)))..)))).----- ( -40.60) >ppk.2978 336200 89 + 6181863 ------------------------UGUCGCGCCCGCGUGCACUGCGGGCGCGAAUGCGGCACAAUUGUGCCCAUUGCGUUG-AACAUGCCCGGGCUGUGAGCCCCGGCCGGAGU------ ------------------------..(((((((((((.....)))))))))))(((.(((((....)))))))).......-.....(((.((((.....)))).)))......------ ( -46.00) >pel.151 303916 90 - 5888780 ------------------------UGUCGCGCCCGCGUGCACUGCGGGCGCGAAUCCGGCACAAUUGCGCUUGUCGCUUUGAAACAUGCCCGGGCCCGACGCCCCGGCCGGAGU------ ------------------------..(((((((((((.....))))))))))).((((((.(((..(((.....))).)))........(.((((.....)))).)))))))..------ ( -45.30) >pf5.3537 400626 106 + 7074893 --CCU-CG----CCUGACGUCAGCCAUCGCGCCCGCGUGCACUGUGGGCGUCAAUACGGCACAAUUGCGCGCAUUCGUUUU-UACAUGCCCGGGCCUGCCGCCCAGGCCGGAGU------ --.((-((----((((.((.(((((...(((((((((.....)))))))))......((((.((..(((......)))..)-)...))))..)).))).))..)))))..))).------ ( -38.70) >consensus ________________________CGUCGCGCCCGCGUGCACUGCGGGCGCCAAUGCGGCACAAUUGCGCUCAUCCCGUUA_AACAUGCCCGGGCUGUCCGCCCCGGCCGGAGU______ ............................(((((((((.....)))))))))......(((........)))................(((.((((.....)))).)))............ (-25.29 = -25.60 + 0.31)


| Location | 406,504 – 406,618 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -47.45 |
| Consensus MFE | -22.23 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
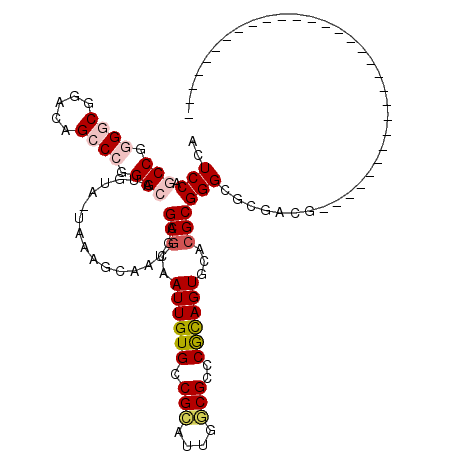
>pao.225 406504 114 + 6264404 ACUCCAACCGCGACGGACAGCCACGGCAUCGG-UAACGGGAAUAGCGGAAUUGUGCCGCAUCGGCGCCCGCAGUGCACGCGGGAC-GCCG----ACGGGAGGCCGGCGGAGAGACGCGGA ..(((..(((..((.((..((....)).)).)-)..))).....(((...((...((((.(((((((((((.......))))).)-))))----)(((....))))))).))..)))))) ( -51.50) >pau.231 421256 112 + 6537648 ACUCCAACCGCGACGGACAGCCACGGCAUCGG-UAACGGGAAUAGCGGAAUUGUGCCGCAUCGGCGCCCGCAGUGCACGCGGGGC-GCCG----ACGGGAGGCCGGCGAAGAGACGCG-- .((((..(((..((.((..((....)).)).)-)..))).....((((.......)))).(((((((((((.......)).))))-))))----)..))))....(((......))).-- ( -48.20) >pfo.3157 429188 84 - 6438405 ACUCCAGCCGGGGCGGGGAGCCCGGGCGC-UA-UGAAUCAAUGCGCGCAAUUGUGCCGCAUUGGCGCCCGUAGUGCACGCGGGCGC---------------------------------- ......(((.((((.....)))).)))..-..-.....((((((((((....))).)))))))((((((((.......))))))))---------------------------------- ( -44.80) >ppk.2978 336200 89 - 6181863 ACUCCGGCCGGGGCUCACAGCCCGGGCAUGUU-CAACGCAAUGGGCACAAUUGUGCCGCAUUCGCGCCCGCAGUGCACGCGGGCGCGACA------------------------------ ......(((.((((.....)))).))).....-.......((((((((....))))).)))((((((((((.......))))))))))..------------------------------ ( -47.90) >pel.151 303916 90 + 5888780 ACUCCGGCCGGGGCGUCGGGCCCGGGCAUGUUUCAAAGCGACAAGCGCAAUUGUGCCGGAUUCGCGCCCGCAGUGCACGCGGGCGCGACA------------------------------ ..(((((((.((((.....)))).))).................((((....)))))))).((((((((((.......))))))))))..------------------------------ ( -47.80) >pf5.3537 400626 106 - 7074893 ACUCCGGCCUGGGCGGCAGGCCCGGGCAUGUA-AAAACGAAUGCGCGCAAUUGUGCCGUAUUGACGCCCACAGUGCACGCGGGCGCGAUGGCUGACGUCAGG----CG-AGG-------- .(((((((((((((.....)))))))).....-....((..((((.(((.(((((.(((....)))..)))))))).))))..))))..(.(((....))).----))-)).-------- ( -44.50) >consensus ACUCCAGCCGGGGCGGACAGCCCGGGCAUGUA_UAAAGCAAUGAGCGCAAUUGUGCCGCAUUGGCGCCCGCAGUGCACGCGGGCGCGACG______________________________ ..(((.(((.((((.....)))).))).................(((..((((((.(((....)))..))))))...))))))..................................... (-22.23 = -23.07 + 0.84)
| Location | 406,504 – 406,618 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.225 406504 114 - 6264404 UCCGCGUCUCUCCGCCGGCCUCCCGU----CGGC-GUCCCGCGUGCACUGCGGGCGCCGAUGCGGCACAAUUCCGCUAUUCCCGUUA-CCGAUGCCGUGGCUGUCCGUCGCGGUUGGAGU .........((((((((((.((..((----((((-(.((((((.....)))))))))))))((((.......))))...........-..)).)))(((((.....))))))).))))). ( -50.30) >pau.231 421256 112 - 6537648 --CGCGUCUCUUCGCCGGCCUCCCGU----CGGC-GCCCCGCGUGCACUGCGGGCGCCGAUGCGGCACAAUUCCGCUAUUCCCGUUA-CCGAUGCCGUGGCUGUCCGUCGCGGUUGGAGU --.(((......)))..(((...(((----((((-((((.(((.....)))))))))))))).)))...(((((........((...-.))..((((((((.....)))))))).))))) ( -49.00) >pfo.3157 429188 84 + 6438405 ----------------------------------GCGCCCGCGUGCACUACGGGCGCCAAUGCGGCACAAUUGCGCGCAUUGAUUCA-UA-GCGCCCGGGCUCCCCGCCCCGGCUGGAGU ----------------------------------(((((((.........)))))))((((((((((....))).)))))))((((.-..-..(((.((((.....)))).)))..)))) ( -40.50) >ppk.2978 336200 89 + 6181863 ------------------------------UGUCGCGCCCGCGUGCACUGCGGGCGCGAAUGCGGCACAAUUGUGCCCAUUGCGUUG-AACAUGCCCGGGCUGUGAGCCCCGGCCGGAGU ------------------------------..(((((((((((.....)))))))))))(((.(((((....)))))))).......-.....(((.((((.....)))).)))...... ( -46.00) >pel.151 303916 90 - 5888780 ------------------------------UGUCGCGCCCGCGUGCACUGCGGGCGCGAAUCCGGCACAAUUGCGCUUGUCGCUUUGAAACAUGCCCGGGCCCGACGCCCCGGCCGGAGU ------------------------------..(((((((((((.....))))))))))).((((((.(((..(((.....))).)))........(.((((.....)))).))))))).. ( -45.30) >pf5.3537 400626 106 + 7074893 --------CCU-CG----CCUGACGUCAGCCAUCGCGCCCGCGUGCACUGUGGGCGUCAAUACGGCACAAUUGCGCGCAUUCGUUUU-UACAUGCCCGGGCCUGCCGCCCAGGCCGGAGU --------.((-((----((((.((.(((((...(((((((((.....)))))))))......((((.((..(((......)))..)-)...))))..)).))).))..)))))..))). ( -38.70) >consensus ______________________________CGUCGCGCCCGCGUGCACUGCGGGCGCCAAUGCGGCACAAUUGCGCUCAUCCCGUUA_AACAUGCCCGGGCUGUCCGCCCCGGCCGGAGU ..................................(((((((((.....)))))))))......(((........)))................(((.((((.....)))).)))...... (-25.29 = -25.60 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:33 2007