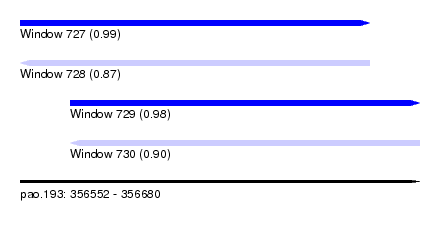
| Sequence ID | pao.193 |
|---|---|
| Location | 356,552 – 356,680 |
| Length | 128 |
| Max. P | 0.986795 |

| Location | 356,552 – 356,664 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.25 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
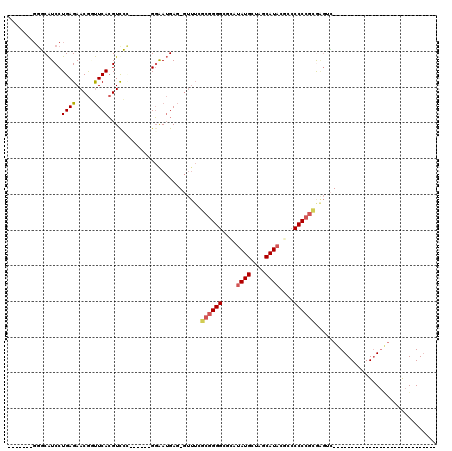
>pao.193 356552 112 + 6264404 GCACCCGCAACUCGGCGCCCAGAGGCAC-UAAGCGCGGGGGGCGUAUGCUAGCAUGUUCUCCCUU-------UUCCUUGCCCGGUUGCGACCUGAGUUGUUCUCAGGACGACCAUGAAUC .....(((((((.((((((....)))..-.(((...((((((..((((....))))..)))))).-------...)))))).))))))).((((((.....))))))............. ( -43.60) >pf5.3563 353482 113 - 7074893 GACACCGCAG--AAAGGCCCUUGGGCACUCUCCUGCGGGGGCGGUAUGCUAGCAUACGCGCCCCGCUAAACACUCAUUCC-----UGCGACGUGAAGCGUUCUCAGGAUGACCAUGAAUU .....(((((--..(((((....))).))...)))))(((((((((((....))))).)))))).........((((.((-----((.((((.....)).)).))))))))......... ( -46.20) >pst.3151 377007 84 - 6397126 -----------------------------GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUUCC------AGAACGUGAACCGUUCUCAGGAUGCCCAUGAAUU -----------------------------...((((((((..((((((....))))))..))))))))...-..(((((.------((((((.....))))))..))))).......... ( -34.10) >psp.2850 373732 74 - 5928787 -----------------------------GACUCACGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUCCC-------GAACGUGAACCGUUCUCAGGAUGC--------- -----------------------------...((.(((((..((((((....))))))..))))).))...-..(((((.-------(((((.....)))))...))))).--------- ( -29.80) >pss.2949 334632 76 - 6093698 -----------------------------GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUCCC-------GAACGUGAACCGUUCUCAGGAUGCCC------- -----------------------------...((((((((..((((((....))))))..))))))))...-..(((((.-------(((((.....)))))...)))))...------- ( -35.20) >pfo.3182 388121 106 - 6438405 GUCACCGCAG--AACCGGCCUUGGGCACUGGCCUGCGGGGUCGGUAUGCUAGCAUAGCCACCCCGCUAAACACUCAUUCC-----UGCGACGUGAAGCGUUCUCAGGAUGACC------- ((((((((((--....((((.........)))).(((((((.((((((....))).)))))))))).............)-----)))((((.....))))....)).)))).------- ( -42.80) >consensus _____________________________GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC_CUCAUUCC______GCAACGUGAACCGUUCUCAGGAUGACC_______ ..................................((((((..((((((....))))))..))))))........(((((.........((((.....))))....))))).......... (-17.66 = -18.14 + 0.47)

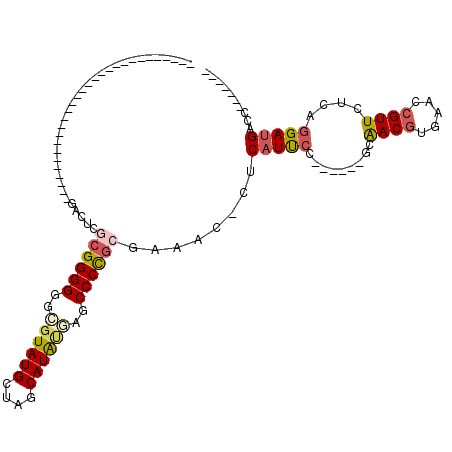

| Location | 356,552 – 356,664 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.25 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.193 356552 112 - 6264404 GAUUCAUGGUCGUCCUGAGAACAACUCAGGUCGCAACCGGGCAAGGAA-------AAGGGAGAACAUGCUAGCAUACGCCCCCCGCGCUUA-GUGCCUCUGGGCGCCGAGUUGCGGGUGC .......(((((.((((((.....)))))).))..)))((((......-------..(......)(((....)))..))))((((((((..-(((((....)))))..)).))))))... ( -45.60) >pf5.3563 353482 113 + 7074893 AAUUCAUGGUCAUCCUGAGAACGCUUCACGUCGCA-----GGAAUGAGUGUUUAGCGGGGCGCGUAUGCUAGCAUACCGCCCCCGCAGGAGAGUGCCCAAGGGCCUUU--CUGCGGUGUC .....(((.((((((((.((.((.....)))).))-----)).)))).)))...((((((((.(((((....)))))))))))(((((.((((.(((....)))))))--)))))))... ( -49.50) >pst.3151 377007 84 + 6397126 AAUUCAUGGGCAUCCUGAGAACGGUUCACGUUCU------GGAAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGCGAGUC----------------------------- ..(((((.....(((..((((((.....))))))------))))))))-..(((((((((..(.((((....)))).)..)))))))))..----------------------------- ( -31.60) >psp.2850 373732 74 + 5928787 ---------GCAUCCUGAGAACGGUUCACGUUC-------GGGAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGUGAGUC----------------------------- ---------.((((((((..(((.....)))))-------))))))..-..(((((((((..(.((((....)))).)..)))))))))..----------------------------- ( -32.40) >pss.2949 334632 76 + 6093698 -------GGGCAUCCUGAGAACGGUUCACGUUC-------GGGAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGCGAGUC----------------------------- -------...((((((((..(((.....)))))-------))))))..-..(((((((((..(.((((....)))).)..)))))))))..----------------------------- ( -34.30) >pfo.3182 388121 106 + 6438405 -------GGUCAUCCUGAGAACGCUUCACGUCGCA-----GGAAUGAGUGUUUAGCGGGGUGGCUAUGCUAGCAUACCGACCCCGCAGGCCAGUGCCCAAGGCCGGUU--CUGCGGUGAC -------.((((((....((.((.....))))(((-----(((...........(((((((((.((((....)))))).))))))).((((.........))))..))--)))))))))) ( -43.10) >consensus _______GGGCAUCCUGAGAACGGUUCACGUCCC______GGAAUGAG_GUUUCGCGGGGCGCAUAUGCUAGCAUACGCCCCCCGCGAGUC_____________________________ ...............((((.....))))..........................((((((....((((....))))....)))))).................................. (-13.36 = -13.78 + 0.42)
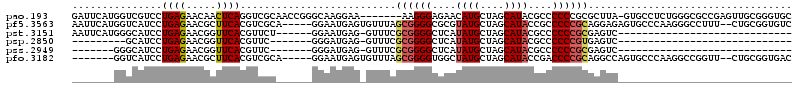
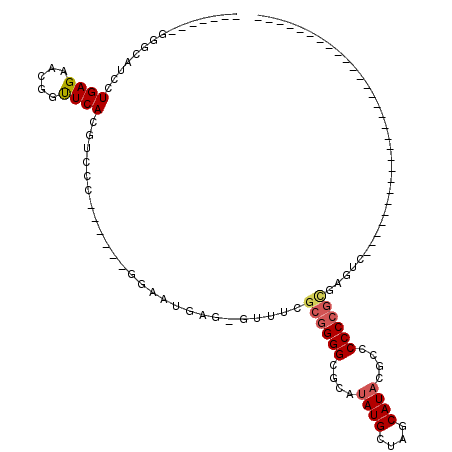
| Location | 356,568 – 356,680 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.14 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
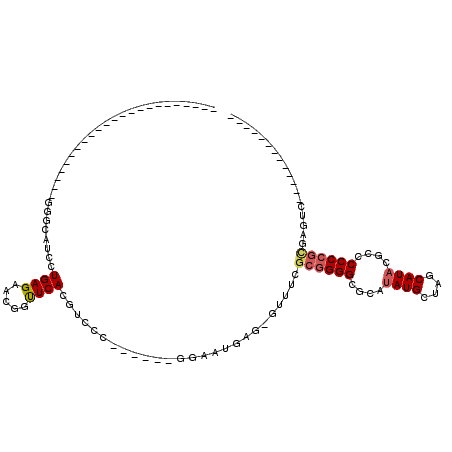
>pao.193 356568 112 + 6264404 GCCCAGAGGCAC-UAAGCGCGGGGGGCGUAUGCUAGCAUGUUCUCCCUU-------UUCCUUGCCCGGUUGCGACCUGAGUUGUUCUCAGGACGACCAUGAAUCAUUUGAGAGUUCCCGA ...((((((((.-.......((((((..((((....))))..)))))).-------.....)))).(((((...((((((.....)))))).)))))........))))........... ( -34.14) >pf5.3563 353496 111 - 7074893 GCCCUUGGGCACUCUCCUGCGGGGGCGGUAUGCUAGCAUACGCGCCCCGCUAAACACUCAUUCC-----UGCGACGUGAAGCGUUCUCAGGAUGACCAUGAAUUGUUCGAGAGUUC---- (((....)))((((((..((((((((((((((....))))).)))))).........((((.((-----((.((((.....)).)).))))))))........)))..))))))..---- ( -47.20) >pst.3151 377007 95 - 6397126 -------------GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUUCC------AGAACGUGAACCGUUCUCAGGAUGCCCAUGAAUUGUCCGAGAGUU----- -------------...((((((((..((((((....))))))..))))))))(((-(((.....------((((((.....))))))..((((...........))))))).)))----- ( -37.90) >psp.2850 373732 74 - 5928787 -------------GACUCACGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUCCC-------GAACGUGAACCGUUCUCAGGAUGC------------------------- -------------...((.(((((..((((((....))))))..))))).))...-..(((((.-------(((((.....)))))...))))).------------------------- ( -29.80) >pss.2949 334632 76 - 6093698 -------------GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC-CUCAUCCC-------GAACGUGAACCGUUCUCAGGAUGCCC----------------------- -------------...((((((((..((((((....))))))..))))))))...-..(((((.-------(((((.....)))))...)))))...----------------------- ( -35.20) >pfo.3182 388135 92 - 6438405 GGCCUUGGGCACUGGCCUGCGGGGUCGGUAUGCUAGCAUAGCCACCCCGCUAAACACUCAUUCC-----UGCGACGUGAAGCGUUCUCAGGAUGACC----------------------- ((((.........)))).(((((((.((((((....))).)))))))))).......((((.((-----((.((((.....)).)).))))))))..----------------------- ( -38.80) >consensus _____________GACUCGCGGGGGGCGUAUGCUAGCAUAUGAGCCCCGCGAAAC_CUCAUUCC______GCAACGUGAACCGUUCUCAGGAUGACC_______________________ ..................((((((..((((((....))))))..))))))........(((((.........((((.....))))....))))).......................... (-17.66 = -18.14 + 0.47)



| Location | 356,568 – 356,680 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.14 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
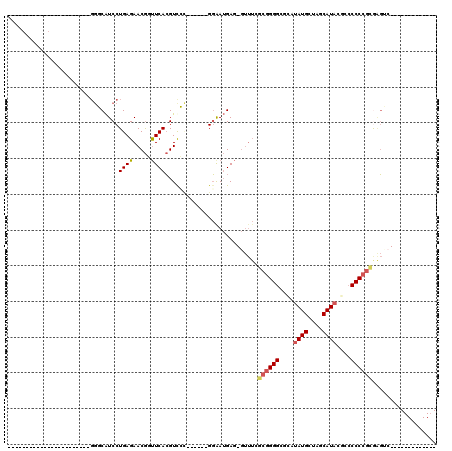
>pao.193 356568 112 - 6264404 UCGGGAACUCUCAAAUGAUUCAUGGUCGUCCUGAGAACAACUCAGGUCGCAACCGGGCAAGGAA-------AAGGGAGAACAUGCUAGCAUACGCCCCCCGCGCUUA-GUGCCUCUGGGC .((((..................(((((.((((((.....)))))).))..)))((((......-------..(......)(((....)))..)))))))).(((((-(.....)))))) ( -34.00) >pf5.3563 353496 111 + 7074893 ----GAACUCUCGAACAAUUCAUGGUCAUCCUGAGAACGCUUCACGUCGCA-----GGAAUGAGUGUUUAGCGGGGCGCGUAUGCUAGCAUACCGCCCCCGCAGGAGAGUGCCCAAGGGC ----..(((((((((((.(((((.....(((((.((.((.....)))).))-----))))))))))))).((((((((.(((((....))))))).))))))..))))))(((....))) ( -49.90) >pst.3151 377007 95 + 6397126 -----AACUCUCGGACAAUUCAUGGGCAUCCUGAGAACGGUUCACGUUCU------GGAAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGCGAGUC------------- -----(((.((((..((..(((.((....)))))(((((.....))))))------)...))))-)))((((((((..(.((((....)))).)..))))))))...------------- ( -33.70) >psp.2850 373732 74 + 5928787 -------------------------GCAUCCUGAGAACGGUUCACGUUC-------GGGAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGUGAGUC------------- -------------------------.((((((((..(((.....)))))-------))))))..-..(((((((((..(.((((....)))).)..)))))))))..------------- ( -32.40) >pss.2949 334632 76 + 6093698 -----------------------GGGCAUCCUGAGAACGGUUCACGUUC-------GGGAUGAG-GUUUCGCGGGGCUCAUAUGCUAGCAUACGCCCCCCGCGAGUC------------- -----------------------...((((((((..(((.....)))))-------))))))..-..(((((((((..(.((((....)))).)..)))))))))..------------- ( -34.30) >pfo.3182 388135 92 + 6438405 -----------------------GGUCAUCCUGAGAACGCUUCACGUCGCA-----GGAAUGAGUGUUUAGCGGGGUGGCUAUGCUAGCAUACCGACCCCGCAGGCCAGUGCCCAAGGCC -----------------------((((.(((((.((.((.....)))).))-----)))...........(((((((((.((((....)))))).))))))).))))...(((...))). ( -38.50) >consensus _______________________GGGCAUCCUGAGAACGGUUCACGUCCC______GGAAUGAG_GUUUCGCGGGGCGCAUAUGCUAGCAUACGCCCCCCGCGAGUC_____________ ...............................((((.....))))..........................((((((....((((....))))....)))))).................. (-13.36 = -13.78 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:27 2007