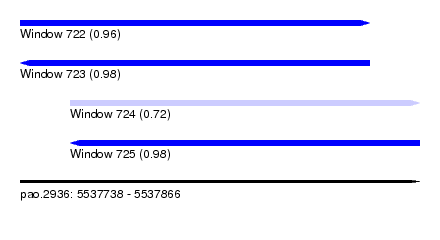
| Sequence ID | pao.2936 |
|---|---|
| Location | 5,537,738 – 5,537,866 |
| Length | 128 |
| Max. P | 0.984686 |

| Location | 5,537,738 – 5,537,850 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -48.63 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.17 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
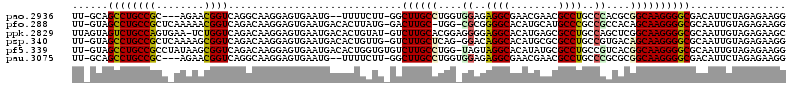
>pao.2936 5537738 112 + 6264404 AAAUGCUCCUUACGGGUU-GCAGCCUGCCGC---AGAACGGUCAGGCAAGGAGUGAAUG--UUUUCUU-GGCUUGCCUGGUGGAGAGGCGAACGAACGCCUGCCCACGCGGCAAGGGGC ....(((((((..((((.-...))))(((((---........((((((((....(((..--..)))..-..))))))))((((..(((((......)))))..)))))))))))))))) ( -53.80) >pfo.288 5816639 113 - 6438405 --AUGCUCCUUACGGGUU-GUAGCCUGCCGCUCAAAAACGGUCAGACAAGGAGUGAAUGACACUUAUG-GACUUGC-UGG-CGCGGGGCACAUGCAUGCCCGCCGCCACAGCAAGGGGC --..(((((((....(((-(((((((((((........))).))).(...(((((.....)))))...-)....))-)((-((.((((((......))))).))))))))))))))))) ( -41.90) >ppk.2829 5544859 115 + 6181863 --AUGCUCCUUACGGGUUAGUAGUCUGCCAGUGAA-UCUGGUCAGACAAGGAGUGAAUGACACUGUAU-GUCUUGCACGGAGGGGAGGCACAUGAGCGCCUGCCAGCUCGGCAAGGGGC --..(((((((.((((((....(((((((((....-.)))).))))).....(((...((((.....)-)))...)))....((.((((.(....).)))).))))))))..))))))) ( -48.50) >psp.340 5251467 114 - 5928787 --AUGCUCCUUACGGGUU-GUAGCCUGCCGCUCAAAAGCGGUCAGACAAGGAGUGAAUGACACUGUUG-GUCUUGCUCAG-GGACAGGCACAUGCGCGCCUGCCGUGACAGCAAGGGGC --..(((((((.(((((.-...)))))(((((....)))))..((((.((.((((.....)))).)).-)))).(((((.-((.(((((.(....).))))))).)))..))))))))) ( -46.00) >pf5.339 6399905 115 - 7074893 --AUGCUCCUUACGGGUU-GUAGCCUGCCGCCUAUAAGCGGUCAGACAAGGAGUGAAUGACACUGGUGUGUCUUGCCUGG-UAGUAGGCACAUAUGCGCCUGCCGUCACGGCAAGGGGC --..(((((((.(((((.-...)))))((((......))))......)))))))....(((((....)))))((((((((-..((((((.(....).))))))..))).)))))..... ( -48.20) >pau.3075 5809116 110 + 6537648 --AUGCUCCUUACGGGUU-GCAGCCUGCCGC---AGAACGGUCAGGCAAGGAGUGAAUG--UUUUCUU-GGCUUGCCUGGUGGAGAGGCGAACGAACGCCUGCCCGCGCGGCAAGGGGC --..(((((((..((((.-...))))(((((---........((((((((....(((..--..)))..-..))))))))((((..(((((......)))))..)))))))))))))))) ( -53.40) >consensus __AUGCUCCUUACGGGUU_GUAGCCUGCCGCU_AAAAACGGUCAGACAAGGAGUGAAUGACACUGCUG_GUCUUGCCUGG_GGAGAGGCACAUGAACGCCUGCCGGCACGGCAAGGGGC ....(((((((.(((((.....)))))(((........)))......))))))).................((((((....((..((((........))))..))....)))))).... (-30.44 = -30.17 + -0.28)


| Location | 5,537,738 – 5,537,850 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -46.63 |
| Consensus MFE | -37.74 |
| Energy contribution | -36.08 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2936 5537738 112 - 6264404 GCCCCUUGCCGCGUGGGCAGGCGUUCGUUCGCCUCUCCACCAGGCAAGCC-AAGAAAA--CAUUCACUCCUUGCCUGACCGUUCU---GCGGCAGGCUGC-AACCCGUAAGGAGCAUUU ((..(((((((((((((.(((((......))))).)))))((((((((..-..(((..--..)))....))))))))........---))))))))..))-...((....))....... ( -50.50) >pfo.288 5816639 113 + 6438405 GCCCCUUGCUGUGGCGGCGGGCAUGCAUGUGCCCCGCG-CCA-GCAAGUC-CAUAAGUGUCAUUCACUCCUUGUCUGACCGUUUUUGAGCGGCAGGCUAC-AACCCGUAAGGAGCAU-- ((.((((((((((((((.((((((....))))))).))-)))-.))....-....((((.....))))....(((((.(((((....))))))))))...-.....)))))).))..-- ( -45.70) >ppk.2829 5544859 115 - 6181863 GCCCCUUGCCGAGCUGGCAGGCGCUCAUGUGCCUCCCCUCCGUGCAAGAC-AUACAGUGUCAUUCACUCCUUGUCUGACCAGA-UUCACUGGCAGACUACUAACCCGUAAGGAGCAU-- ((.((((((.(((..((.((((((....)))))))).))).(((...(((-((...)))))...))).....(((((.((((.-....))))))))).........)))))).))..-- ( -43.40) >psp.340 5251467 114 + 5928787 GCCCCUUGCUGUCACGGCAGGCGCGCAUGUGCCUGUCC-CUGAGCAAGAC-CAACAGUGUCAUUCACUCCUUGUCUGACCGCUUUUGAGCGGCAGGCUAC-AACCCGUAAGGAGCAU-- ((.(((((((((((.(((((((((....))))))).))-.))).))....-....((((.....))))....(((((.(((((....))))))))))...-.....)))))).))..-- ( -45.90) >pf5.339 6399905 115 + 7074893 GCCCCUUGCCGUGACGGCAGGCGCAUAUGUGCCUACUA-CCAGGCAAGACACACCAGUGUCAUUCACUCCUUGUCUGACCGCUUAUAGGCGGCAGGCUAC-AACCCGUAAGGAGCAU-- (((....((((...))))((((((....))))))....-...)))..(((((....))))).....(((((((((((.(((((....)))))))))).((-.....))))))))...-- ( -47.20) >pau.3075 5809116 110 - 6537648 GCCCCUUGCCGCGCGGGCAGGCGUUCGUUCGCCUCUCCACCAGGCAAGCC-AAGAAAA--CAUUCACUCCUUGCCUGACCGUUCU---GCGGCAGGCUGC-AACCCGUAAGGAGCAU-- ((..((((((((..(((.(((((......))))).)))..((((((((..-..(((..--..)))....))))))))........---))))))))..))-...((....)).....-- ( -47.10) >consensus GCCCCUUGCCGUGACGGCAGGCGCUCAUGUGCCUCUCC_CCAGGCAAGAC_AAAAAGUGUCAUUCACUCCUUGUCUGACCGUUUUU_AGCGGCAGGCUAC_AACCCGUAAGGAGCAU__ ....((((((.....((.((((((....)))))).)).....))))))..................(((((((((((.((((......))))))))).((......))))))))..... (-37.74 = -36.08 + -1.66)

| Location | 5,537,754 – 5,537,866 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -44.57 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.05 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
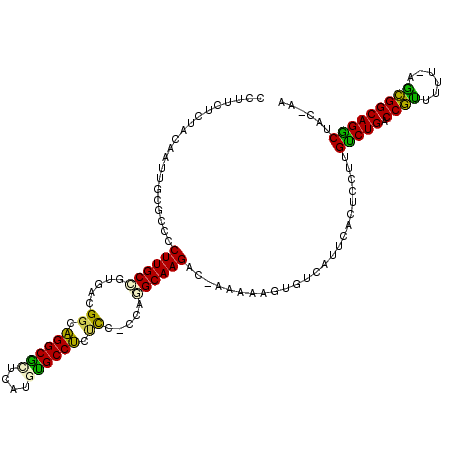
>pao.2936 5537754 112 + 6264404 UU-GCAGCCUGCCGC---AGAACGGUCAGGCAAGGAGUGAAUG--UUUUCUU-GGCUUGCCUGGUGGAGAGGCGAACGAACGCCUGCCCACGCGGCAAGGGGCGACAUUCUAGAGAAGG ((-((..((((((((---........((((((((....(((..--..)))..-..))))))))((((..(((((......)))))..))))))))).))).)))).............. ( -48.50) >pfo.288 5816653 115 - 6438405 UU-GUAGCCUGCCGCUCAAAAACGGUCAGACAAGGAGUGAAUGACACUUAUG-GACUUGC-UGG-CGCGGGGCACAUGCAUGCCCGCCGCCACAGCAAGGGGCGCAAUUGUAGAGAAGG ((-((.((((((((........))))....(...(((((.....)))))...-).(((((-(((-((.((((((......))))).)))))..))))))))))))))............ ( -39.40) >ppk.2829 5544873 117 + 6181863 UUAGUAGUCUGCCAGUGAA-UCUGGUCAGACAAGGAGUGAAUGACACUGUAU-GUCUUGCACGGAGGGGAGGCACAUGAGCGCCUGCCAGCUCGGCAAGGGGCGCAAUUGUAGAGAAGC ......(((((((((....-.)))).)))))....((((.....))))(..(-.((((((.(((..((.((((.(....).)))).))..).)))))))).)..).............. ( -41.10) >psp.340 5251481 116 - 5928787 UU-GUAGCCUGCCGCUCAAAAGCGGUCAGACAAGGAGUGAAUGACACUGUUG-GUCUUGCUCAG-GGACAGGCACAUGCGCGCCUGCCGUGACAGCAAGGGGCGCAAUUGUAGAGAAGG .(-((.((((((((((....)))(((.((((.((.((((.....)))).)).-)))).)))...-)).))))))))(((((.(((((.......)).))).)))))............. ( -43.00) >pf5.339 6399919 117 - 7074893 UU-GUAGCCUGCCGCCUAUAAGCGGUCAGACAAGGAGUGAAUGACACUGGUGUGUCUUGCCUGG-UAGUAGGCACAUAUGCGCCUGCCGUCACGGCAAGGGGCGCAAUUGUAGAGAAGG ..-.....(((((((......)))).)))((((..((((.....)))).((((.((((((((((-..((((((.(....).))))))..))).))))))).))))..))))........ ( -47.30) >pau.3075 5809130 112 + 6537648 UU-GCAGCCUGCCGC---AGAACGGUCAGGCAAGGAGUGAAUG--UUUUCUU-GGCUUGCCUGGUGGAGAGGCGAACGAACGCCUGCCCGCGCGGCAAGGGGCGACAUUCUAGAGAAGG ((-((..((((((((---........((((((((....(((..--..)))..-..))))))))((((..(((((......)))))..))))))))).))).)))).............. ( -48.10) >consensus UU_GUAGCCUGCCGCU_AAAAACGGUCAGACAAGGAGUGAAUGACACUGCUG_GUCUUGCCUGG_GGAGAGGCACAUGAACGCCUGCCGGCACGGCAAGGGGCGCAAUUGUAGAGAAGG ......((((((((........)))).............................((((((....((..((((........))))..))....))))))))))................ (-24.33 = -24.05 + -0.28)
| Location | 5,537,754 – 5,537,866 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -32.49 |
| Energy contribution | -31.05 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.52 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
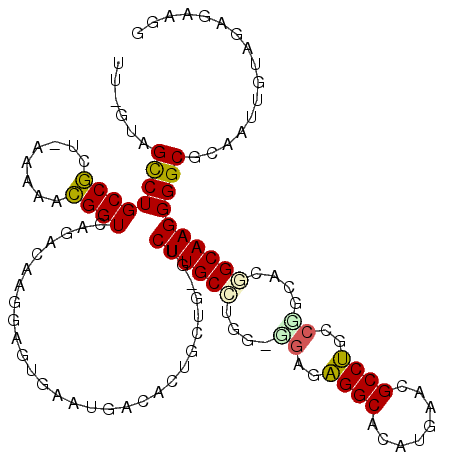
>pao.2936 5537754 112 - 6264404 CCUUCUCUAGAAUGUCGCCCCUUGCCGCGUGGGCAGGCGUUCGUUCGCCUCUCCACCAGGCAAGCC-AAGAAAA--CAUUCACUCCUUGCCUGACCGUUCU---GCGGCAGGCUGC-AA ............(((.(((...(((((((((((.(((((......))))).)))))((((((((..-..(((..--..)))....))))))))........---))))))))).))-). ( -47.70) >pfo.288 5816653 115 + 6438405 CCUUCUCUACAAUUGCGCCCCUUGCUGUGGCGGCGGGCAUGCAUGUGCCCCGCG-CCA-GCAAGUC-CAUAAGUGUCAUUCACUCCUUGUCUGACCGUUUUUGAGCGGCAGGCUAC-AA ....................((((((..(((((.((((((....))))))).))-)))-)))))..-....((((.....))))....(((((.(((((....))))))))))...-.. ( -39.80) >ppk.2829 5544873 117 - 6181863 GCUUCUCUACAAUUGCGCCCCUUGCCGAGCUGGCAGGCGCUCAUGUGCCUCCCCUCCGUGCAAGAC-AUACAGUGUCAUUCACUCCUUGUCUGACCAGA-UUCACUGGCAGACUACUAA ............((((((....((((.....))))(((((....)))))........))))))(((-((...)))))...........(((((.((((.-....)))))))))...... ( -37.00) >psp.340 5251481 116 + 5928787 CCUUCUCUACAAUUGCGCCCCUUGCUGUCACGGCAGGCGCGCAUGUGCCUGUCC-CUGAGCAAGAC-CAACAGUGUCAUUCACUCCUUGUCUGACCGCUUUUGAGCGGCAGGCUAC-AA .............(((((.(((.((((...))))))).)))))((((((((((.-((((((.((((-....((((.....))))....))))....))))...)).))))))).))-). ( -41.40) >pf5.339 6399919 117 + 7074893 CCUUCUCUACAAUUGCGCCCCUUGCCGUGACGGCAGGCGCAUAUGUGCCUACUA-CCAGGCAAGACACACCAGUGUCAUUCACUCCUUGUCUGACCGCUUAUAGGCGGCAGGCUAC-AA ........(((..((((((...(((((...)))))))))))..)))((((....-..))))..(((((....)))))...........(((((.(((((....))))))))))...-.. ( -45.70) >pau.3075 5809130 112 - 6537648 CCUUCUCUAGAAUGUCGCCCCUUGCCGCGCGGGCAGGCGUUCGUUCGCCUCUCCACCAGGCAAGCC-AAGAAAA--CAUUCACUCCUUGCCUGACCGUUCU---GCGGCAGGCUGC-AA ............(((.(((...((((((..(((.(((((......))))).)))..((((((((..-..(((..--..)))....))))))))........---))))))))).))-). ( -44.30) >consensus CCUUCUCUACAAUUGCGCCCCUUGCCGUGACGGCAGGCGCUCAUGUGCCUCUCC_CCAGGCAAGAC_AAAAAGUGUCAUUCACUCCUUGUCUGACCGUUUUU_AGCGGCAGGCUAC_AA ....................((((((.....((.((((((....)))))).)).....))))))........................(((((.((((......)))))))))...... (-32.49 = -31.05 + -1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:23 2007