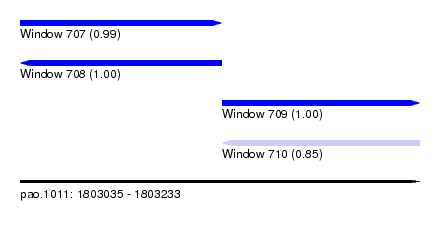
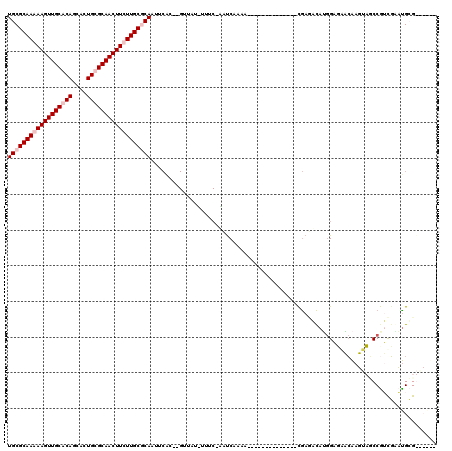
| Sequence ID | pao.1011 |
|---|---|
| Location | 1,803,035 – 1,803,233 |
| Length | 198 |
| Max. P | 0.997941 |

| Location | 1,803,035 – 1,803,135 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.35 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -28.19 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1011 1803035 100 + 6264404 ------CUCAUUCAAUGGCUUAUUGUUAUCCAUGUCUCA--------------UUCUGAAAAGAAAUAUAACAAGUGAAUUGAGCAAGAAGUUGCGCAGUGCUGUGCAACUUUUUGCGCA ------.....(((((..(...(((((((..........--------------((((....))))..)))))))..).)))))(((((((((((((((....)))))))))))))))... ( -32.10) >pau.2014 2705870 99 - 6537648 ------CGCAUCCGACGGAUAAUUGUUAUCCAUGUCUCG--------------UUUUGAAU-GAAAAAUAACAAGUGAAUUGAGCAAGAAGUUGCGCAGUGCUGUGCAACUUUUUGCGCA ------.((.(((((((((((.....)))))..)))..(--------------((((....-..))))).....).)).....(((((((((((((((....))))))))))))))))). ( -32.70) >pfo.1173 2331605 118 + 6438405 CACGUCCCCGGGAGCCGGGGACUCCACCGCUCUGGAUCGAGGCUUUGAGCGUUUUUUGAUCAGCCAGGUAGC--UCGAAGUGCGCAAGAAGUUGCGCAGUACUGCGCAACUUCUUGCGCA ...((((((((...))))))))((((......)))).....(((((((((....((((......))))..))--)))))))(((((((((((((((((....))))))))))))))))). ( -66.40) >pst.3233 216137 95 - 6397126 UACGCCCGCAGUCG-CGGCGACUUGAUCCCCUUACGUUG-----UAAAAAAGCUUUUGGUU-------------------UGCGCAAGAAGUUGCGCACUGUUGCGCAACUUCUUGCGCA ..((((.((....)-))))).........((..(.(((.-----......))).)..))..-------------------((((((((((((((((((....)))))))))))))))))) ( -42.60) >consensus ______CGCAGUCGACGGCGAAUUGAUAUCCAUGUCUCG______________UUUUGAAU_GAAA_AUAAC__GUGAAUUGAGCAAGAAGUUGCGCAGUGCUGCGCAACUUCUUGCGCA ................((.(.......).)).................................................((((((((((((((((((....)))))))))))))))))) (-28.19 = -27.50 + -0.69)

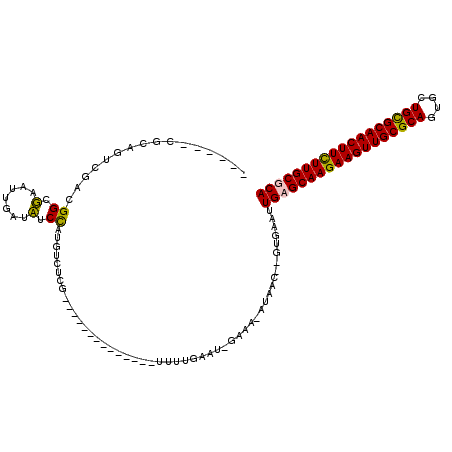
| Location | 1,803,035 – 1,803,135 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.35 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -24.15 |
| Energy contribution | -25.65 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1011 1803035 100 - 6264404 UGCGCAAAAAGUUGCACAGCACUGCGCAACUUCUUGCUCAAUUCACUUGUUAUAUUUCUUUUCAGAA--------------UGAGACAUGGAUAACAAUAAGCCAUUGAAUGAG------ ...((((.(((((((.((....)).))))))).))))(((((....(((((((...((((.......--------------.)))).....)))))))......))))).....------ ( -23.80) >pau.2014 2705870 99 + 6537648 UGCGCAAAAAGUUGCACAGCACUGCGCAACUUCUUGCUCAAUUCACUUGUUAUUUUUC-AUUCAAAA--------------CGAGACAUGGAUAACAAUUAUCCGUCGGAUGCG------ .((((((.(((((((.((....)).))))))).))))...((((.((((((.......-......))--------------))))..(((((((.....))))))).)))))).------ ( -28.62) >pfo.1173 2331605 118 - 6438405 UGCGCAAGAAGUUGCGCAGUACUGCGCAACUUCUUGCGCACUUCGA--GCUACCUGGCUGAUCAAAAAACGCUCAAAGCCUCGAUCCAGAGCGGUGGAGUCCCCGGCUCCCGGGGACGUG ((((((((((((((((((....))))))))))))))))))......--.(((((.((((.................))))((......))..))))).((((((((...))))))))... ( -61.03) >pst.3233 216137 95 + 6397126 UGCGCAAGAAGUUGCGCAACAGUGCGCAACUUCUUGCGCA-------------------AACCAAAAGCUUUUUUA-----CAACGUAAGGGGAUCAAGUCGCCG-CGACUGCGGGCGUA ((((((((((((((((((....))))))))))))))))))-------------------........((.......-----(..(....)..).........(((-(....))))))... ( -43.10) >consensus UGCGCAAAAAGUUGCACAGCACUGCGCAACUUCUUGCGCAAUUCAC__GUUAU_UUUC_AAUCAAAA______________CGAGACAUGGAGAACAAGUAGCCGUCGAAUGCG______ ((((((((((((((((((....))))))))))))))))))................................................................................ (-24.15 = -25.65 + 1.50)

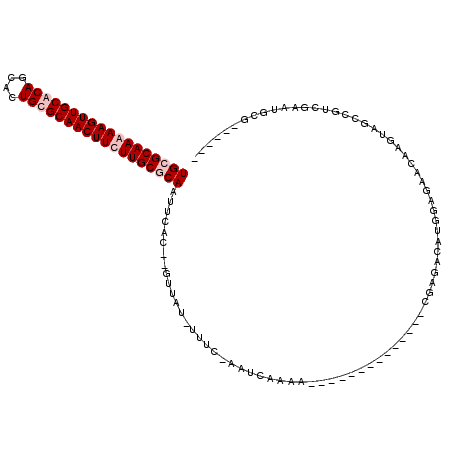
| Location | 1,803,135 – 1,803,233 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 69.08 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -18.00 |
| Energy contribution | -14.47 |
| Covariance contribution | -3.54 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
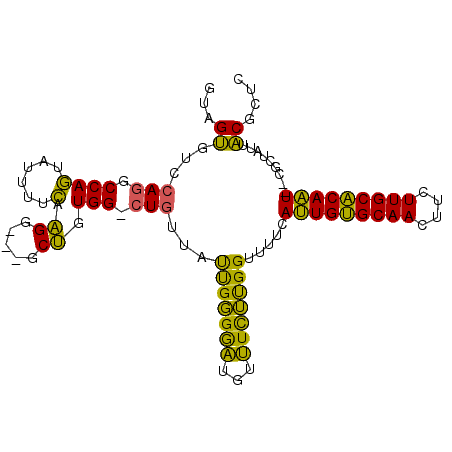
>pao.1011 1803135 98 + 6264404 GUAGUGUUCAGGCCAGUAUUUUCAAGG---GCUGUGGACUGUUAUUGGGGAUGUUUCUUGGUUUUCAUUGUGCAACUUCUUGCACAAU--CGCUAUUACGCUC ((((((...(((((((((((..(((..---((........))..)))..))))....)))))))..(((((((((....)))))))))--))))))....... ( -26.90) >pau.2014 2705969 98 - 6537648 GUAGUGUCCAGGCCAGUAUUUUCAAGG---GCUGUGGGCUGUUAUUGGGGAUGUUUCUUGGUUUUCAUUGUGCAACUUCUUGCACAAU--CGCUAUUACGCUC ((((((.(((((....((((..(((..---((........))..)))..))))...))))).....(((((((((....)))))))))--))))))....... ( -27.40) >pfo.1349 3754958 100 - 6438405 CUUGGCUGUAAGCCACGAUAUAGAGGGCUUGCCGUGG-CUUUCACCGUGAUUUUAUUACGGGC--AACUGCGCAACUUUUUGCGCAGUUCCGCCAAUCCGAUC .(((((((.((((((((....((....))...)))))-))).))(((((((...)))))))(.--((((((((((....)))))))))).))))))....... ( -40.60) >consensus GUAGUGUCCAGGCCAGUAUUUUCAAGG___GCUGUGG_CUGUUAUUGGGGAUGUUUCUUGGUUUUCAUUGUGCAACUUCUUGCACAAU__CGCUAUUACGCUC ...((...(((.((((......).((.....)).))).)))...(((((((...))))))).....(((((((((....))))))))).........)).... (-18.00 = -14.47 + -3.54)

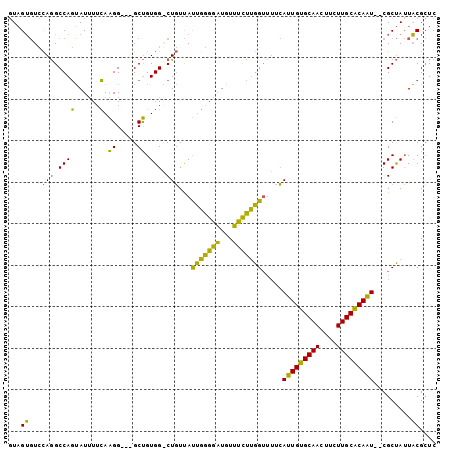
| Location | 1,803,135 – 1,803,233 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.08 |
| Mean single sequence MFE | -24.21 |
| Consensus MFE | -14.59 |
| Energy contribution | -13.38 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
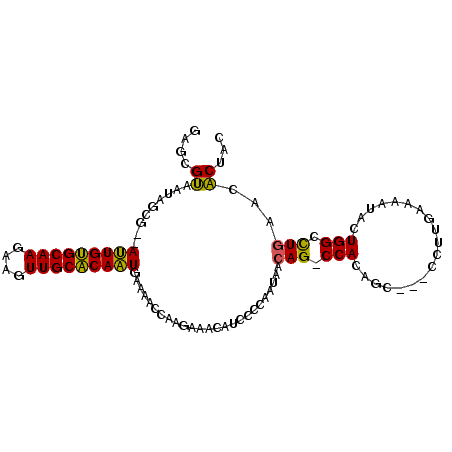
>pao.1011 1803135 98 - 6264404 GAGCGUAAUAGCG--AUUGUGCAAGAAGUUGCACAAUGAAAACCAAGAAACAUCCCCAAUAACAGUCCACAGC---CCUUGAAAAUACUGGCCUGAACACUAC ..((......)).--(((((((((....)))))))))..........................(((...(((.---((...........)).)))...))).. ( -16.70) >pau.2014 2705969 98 + 6537648 GAGCGUAAUAGCG--AUUGUGCAAGAAGUUGCACAAUGAAAACCAAGAAACAUCCCCAAUAACAGCCCACAGC---CCUUGAAAAUACUGGCCUGGACACUAC ..((......)).--(((((((((....)))))))))...........................(.(((..((---(..(....)....))).))).)..... ( -17.00) >pfo.1349 3754958 100 + 6438405 GAUCGGAUUGGCGGAACUGCGCAAAAAGUUGCGCAGUU--GCCCGUAAUAAAAUCACGGUGAAAG-CCACGGCAAGCCCUCUAUAUCGUGGCUUACAGCCAAG .......((((((.((((((((((....))))))))))--.)((((.........))))((.(((-((((((.............))))))))).))))))). ( -38.92) >consensus GAGCGUAAUAGCG__AUUGUGCAAGAAGUUGCACAAUGAAAACCAAGAAACAUCCCCAAUAACAG_CCACAGC___CCUUGAAAAUACUGGCCUGAACACUAC ....((.........(((((((((....))))))))).........................(((.(((...................))).)))...))... (-14.59 = -13.38 + -1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:16:06 2007