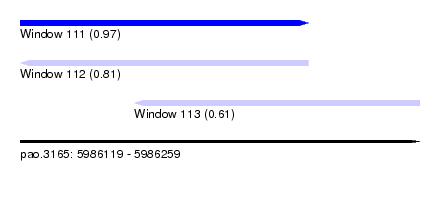
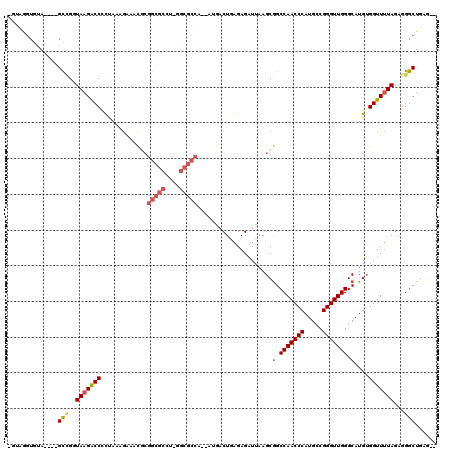
| Sequence ID | pao.3165 |
|---|---|
| Location | 5,986,119 – 5,986,259 |
| Length | 140 |
| Max. P | 0.967689 |

| Location | 5,986,119 – 5,986,220 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -27.13 |
| Energy contribution | -28.63 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
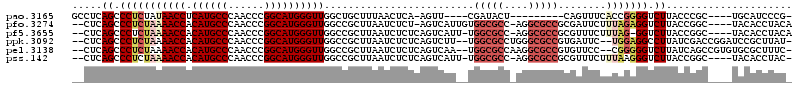
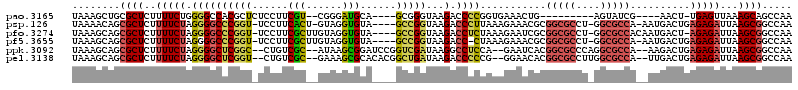
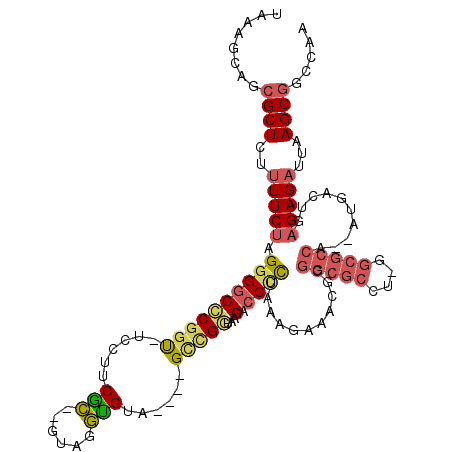
>pao.3165 5986119 101 + 6264404 GCCUCAGCCCUCUAUAACCUCAUGCCCAACCCGGCAUGGGUUGGCUGCUUUAACUCA-AGUU----CGAUACU---------CAGUUUCACCGGGGUCUUACCCGC----UGCAUCCCG- ((..(((((......((((.((((((......)))))))))))))))....((((..-(((.----....)))---------.))))....((((......)))).----.))......- ( -28.30) >pfo.3274 6206311 112 + 6438405 --CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCU-AGUCAUUGUGGCGCC-AGGCGCCGCGAUUCUUUAGAGGUCUUACCGGC----UACACCUACA --.............((((.((((((......))))))))))(((((....((((((-((..((((((((((.-..))))))))))...)))))))).....))))----)......... ( -44.80) >pf5.3655 6871053 111 + 7074893 --CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCUCAGUCAUU-UGGCGCC-AGGCGCCGCGUUUCUUUAG-GGUCUUACCGGC----UACACCUACA --.............((((.((((((......))))))))))(((((......((((.((..((.-((((((.-..)))))).))..))..))-))......))))----)......... ( -34.40) >ppk.3092 6030778 113 + 6181863 --CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCUCAGUCUU--UGGCGCCUGGGCGCCGUGAUUC--UGGAGGCCUUAUCGACCGGAUCCGCUUAU- --...(((..(((..((((.((((((......))))))))))((.((..(((((((.(((....--((((((....)))))).....)--))))))..))).)).)))))...)))...- ( -38.70) >pel.3138 5748237 113 + 5888780 --CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCUCAGUCAA--UGGCGCCAAGGCGCCGUGUUCC--CGGGGGUCUUAUCAGCCGUGUGCGCUUUC- --....((.(.(...((((.((((((......))))))))))(((........(((((.(...(--((((((....)))))))...).--.)))))........)))).).))......- ( -39.19) >pss.142 5854490 111 - 6093698 --CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCUCAGUCAUU-UGGCGCC-AGGCGCCGCGUUUCUUUAAGGGUCUUACCGGC----UACACCUAC- --...((((......((((.((((((......))))))))))((((.(((((......((..((.-((((((.-..)))))).))..))))))))))).....)))----)........- ( -36.40) >consensus __CUCAGCCCUCUAAAACCACAUGCCCAACCCGGCAUGGGUUGGCCGCUUAAUCUCUCAGUCAU__UGGCGCC_AGGCGCCGCGUUUCUUUAGGGGUCUUACCGGC____UACACCUAC_ .....((.(((((((((((.((((((......)))))))))).........................(((((....)))))........))))))).))..................... (-27.13 = -28.63 + 1.50)
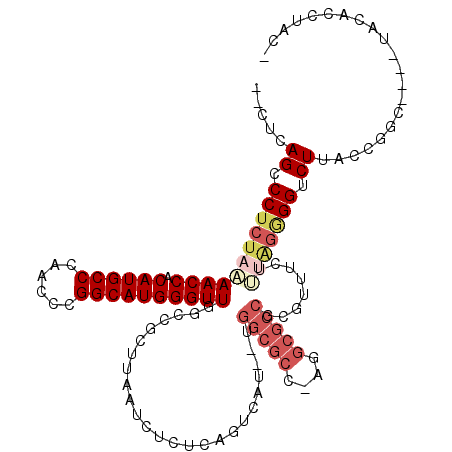
| Location | 5,986,119 – 5,986,220 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -31.32 |
| Energy contribution | -32.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
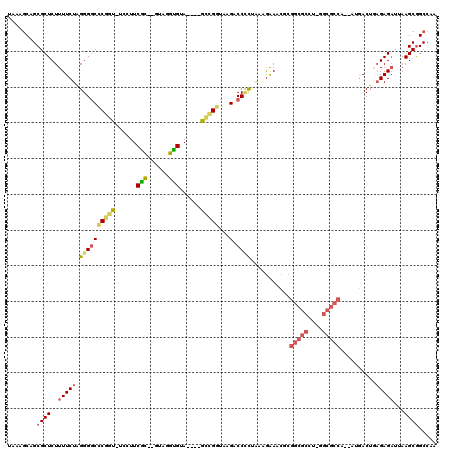
>pao.3165 5986119 101 - 6264404 -CGGGAUGCA----GCGGGUAAGACCCCGGUGAAACUG---------AGUAUCG----AACU-UGAGUUAAAGCAGCCAACCCAUGCCGGGUUGGGCAUGAGGUUAUAGAGGGCUGAGGC -.......((----.((((......)))).)).(((((---------(((....----.)))-).))))....(((((((((((((((......)))))).))))......))))).... ( -33.80) >pfo.3274 6206311 112 - 6438405 UGUAGGUGUA----GCCGGUAAGACCUCUAAAGAAUCGCGGCGCCU-GGCGCCACAAUGACU-AGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG-- ....((....----.))....((.((((((..((((((((((((..-.))))).........-...........(.(((((((.....))))))).)..))))))))))))).))...-- ( -41.40) >pf5.3655 6871053 111 - 7074893 UGUAGGUGUA----GCCGGUAAGACC-CUAAAGAAACGCGGCGCCU-GGCGCCA-AAUGACUGAGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG-- ........((----(((((.....))-((((((..(((((((((..-.))))).-..(((.(....).))).))(.(((((((.....))))))).)..))..))))))..)))))..-- ( -39.60) >ppk.3092 6030778 113 - 6181863 -AUAAGCGGAUCCGGUCGAUAAGGCCUCCA--GAAUCACGGCGCCCAGGCGCCA--AAGACUGAGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG-- -.....(((.(((((((.....))))((.(--((((((((((((....))))).--..................(.(((((((.....))))))).)..)))))))).))))))))..-- ( -46.70) >pel.3138 5748237 113 - 5888780 -GAAAGCGCACACGGCUGAUAAGACCCCCG--GGAACACGGCGCCUUGGCGCCA--UUGACUGAGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG-- -...(((.(....(((((......((....--)).....(((((....))))).--.................)))))((((((((((......)))))).)))).....).)))...-- ( -41.90) >pss.142 5854490 111 + 6093698 -GUAGGUGUA----GCCGGUAAGACCCUUAAAGAAACGCGGCGCCU-GGCGCCA-AAUGACUGAGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG-- -.......((----(((..(((((((...........(((((((..-.))))).-..(((.(....).))).))(.(((((((.....))))))).)....)))))))...)))))..-- ( -38.60) >consensus _GUAGGUGUA____GCCGGUAAGACCCCUAAAGAAACGCGGCGCCU_GGCGCCA__AUGACUGAGAGAUUAAGCGGCCAACCCAUGCCGGGUUGGGCAUGUGGUUUUAGAGGGCUGAG__ ..............(((..(((((((.............(((((....))))).....................(.(((((((.....))))))).)....)))))))...)))...... (-31.32 = -32.27 + 0.95)


| Location | 5,986,159 – 5,986,259 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3165 5986159 100 - 6264404 UAAAGCUGCGCUCUUUUCUGGGGCCACGCUCUCCUUCGU--CGGGAUGCA----GCGGGUAAGACCCCGGUGAAACUG---------AGUAUCG----AACU-UGAGUUAAAGCAGCCAA ....((((((((((.....)))))...((((...(((((--(((....((----.((((......)))).))...)))---------)....))----))..-.))))....)))))... ( -34.30) >psp.126 5678292 112 + 5928787 UAAAACAGCGCUCUUUUCUAGGGGCCCGGU-UCCUUCACU-GUAGGUGUA----GCCGGUAAGACCCUUAAAGAAACGCGGCGCCU-GGCGCCA-AAUGACUGAGAGAUUAAGCGGCCAA .......((..(((((...((((..(((((-(....(((.-....))).)----))))).....)))).)))))..((((((((..-.))))).-..(((.(....).))).)))))... ( -33.50) >pfo.3274 6206349 113 - 6438405 UAAAGCAGCGCUCUUUUCUAGGGGCCCGGU-UCCUUCGCUUGUAGGUGUA----GCCGGUAAGACCUCUAAAGAAUCGCGGCGCCU-GGCGCCACAAUGACU-AGAGAUUAAGCGGCCAA ....((.((.((((..((((((((((((((-(....((((....)))).)----)))))...).)))))).......(.(((((..-.))))).)...))..-)))).....)).))... ( -35.70) >pf5.3655 6871091 112 - 7074893 UAAAGCAGCGCUCUUUUCUAGGGGCCCGGU-UCCUUCGCUUGUAGGUGUA----GCCGGUAAGACC-CUAAAGAAACGCGGCGCCU-GGCGCCA-AAUGACUGAGAGAUUAAGCGGCCAA ....((.((.(((((.(((((((..(((((-(....((((....)))).)----))))).....))-))).........(((((..-.))))).-...))..))))).....)).))... ( -36.10) >ppk.3092 6030816 112 - 6181863 UAAAGCAGCGCUCUUUUCUAGGGGCUCGGC--CUGUCGC--AUAAGCGGAUCCGGUCGAUAAGGCCUCCA--GAAUCACGGCGCCCAGGCGCCA--AAGACUGAGAGAUUAAGCGGCCAA ....((.((.(((((((((.((((((((((--(..((((--....))))....))))))....))))).)--)))....(((((....))))).--......))))).....)).))... ( -44.00) >pel.3138 5748275 112 - 5888780 UAAAGCAGCGCUCUUUUCUAGGGGCUCGGU--CUGUCGC--GAAAGCGCACACGGCUGAUAAGACCCCCG--GGAACACGGCGCCUUGGCGCCA--UUGACUGAGAGAUUAAGCGGCCAA ....((.((.((((((((..((((((((((--(((((((--....))).))).))))))...).))))..--)))....(((((....))))).--......))))).....)).))... ( -47.60) >consensus UAAAGCAGCGCUCUUUUCUAGGGGCCCGGU_UCCUUCGC__GUAGGUGUA____GCCGGUAAGACCCCUAAAGAAACGCGGCGCCU_GGCGCCA__AUGACUGAGAGAUUAAGCGGCCAA ........((((..(((((.((((((((((......(((......)))......)))))...).))))...........(((((....)))))..........)))))...))))..... (-25.38 = -25.92 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:19 2007