
| Sequence ID | pao.2779 |
|---|---|
| Location | 5,266,896 – 5,267,463 |
| Length | 567 |
| Max. P | 0.999999 |

| Location | 5,266,896 – 5,267,016 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -40.02 |
| Energy contribution | -35.88 |
| Covariance contribution | -4.13 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.08 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930798 |
| Prediction | RNA |
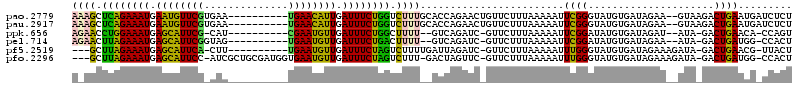
Download alignment: ClustalW | MAF
>pao.2779 5266896 120 - 6264404 CUUAGUAGUGGCGAGCGAACGGGGAUUAGCCCUUAAGCUUCAUUGAUUUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAGGAUCUUUGAAGUGAA (((...((......(((.((((((....((((....(((...........)))((..(((.((....)).))).)).....)))).....)))))).)))........))...))).... ( -36.24) >pau.2917 5538828 120 - 6537648 CUUAGUAGUGGCGAGCGAACGGGGAUUAGCCCUUAAGCUUCAUUGAUUUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAGGAUCUUUGAAGUGAA (((...((......(((.((((((....((((....(((...........)))((..(((.((....)).))).)).....)))).....)))))).)))........))...))).... ( -36.24) >ppk.656 4854192 120 + 6181863 CUUAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGUUGAUUUGAGAUUAGUGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACACGAAAAUCUCUUGUCAAUGAA ........(((((((.((((((((....((((......(((((...)))))((((..(((.((....)).))).))))...)))).....)))))).........)).)))))))..... ( -36.90) >pel.714 4510860 120 + 5888780 CUUAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGCUGGUUUGAGAUUAGUGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACACGAAAAUCUCUUGCCAGUGAA .....((.((((((........((((((((......))))))))((((((.((((..(((.((....)).)))..)...(.((((....)))))...)))...)))))))))))).)).. ( -43.30) >pf5.2519 4812766 120 - 7074893 CUUAGUAGUGGCGAGCGAACGGGGACUAGCCCUUAAGUGGCUUUGAGAUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCUGUACGCGAAAAUCUCUUAGUCAUGAA .......(((((..(((.((((((.(((((((....(((((((((.......)))).(((.((....)).))))))))...))))..))))))))).)))((.....))...)))))... ( -41.70) >pfo.2296 4419329 120 - 6438405 CUUAGUAGUGGCGAGCGAACGGGGACUAGCCCUUAAGUGGCUUUGAGAUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCUGUACGCGAAAGUCUCUUAGUCAUGAA .......(((((..(((.((((((.(((((((....(((((((((.......)))).(((.((....)).))))))))...))))..))))))))).)))((.....))...)))))... ( -41.70) >consensus CUUAGUAGUGGCGAGCGAACGGGGACUAGCCCUUAAGCUGCUUUGAGAUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAAUCUCUUAGAAAUGAA ....................((((.....))))...((((((..((((((.((((..(((.((....)).)))..).....((((....))))....)))...))))))..))))))... (-40.02 = -35.88 + -4.13)

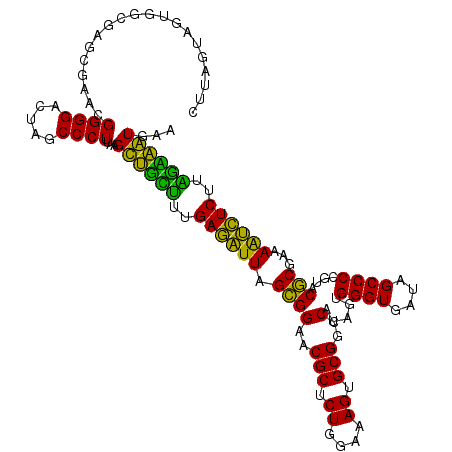
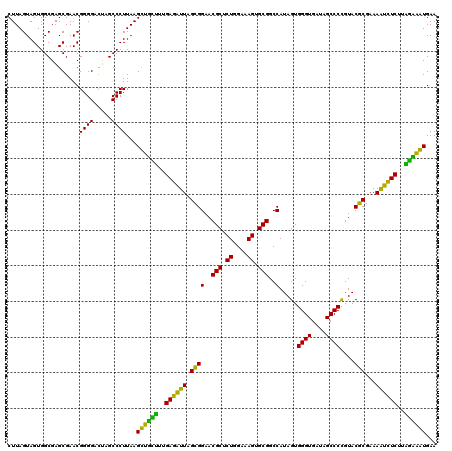
| Location | 5,267,016 – 5,267,136 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -29.66 |
| Energy contribution | -28.33 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267016 120 + 6264404 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCAUU ((((((......))))))...(((..((((((...((.((..((((.....))))..)).))..))).)))..))).............(((((((....)))))))(((....)))... ( -32.30) >pau.2917 5538948 120 + 6537648 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCAUU ((((((......))))))...(((..((((((...((.((..((((.....))))..)).))..))).)))..))).............(((((((....)))))))(((....)))... ( -32.30) >ppk.656 4854312 120 - 6181863 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGCUGGCUGGGUUCCCCCAUU ((((((......)))))).........(((.((((((..(..((((.....))))..)...(((((.((...........)).)))))...(((((....))))))))))).)))..... ( -29.90) >pel.714 4510980 120 - 5888780 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGCUGGCUGGGUUCCCCCAUU ((((((......)))))).........(((.((((((..(..((((.....))))..)...(((((.((...........)).)))))...(((((....))))))))))).)))..... ( -29.90) >pf5.2519 4812886 120 + 7074893 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUU ((((((......)))))).........(((.((((((..(..((((.....))))..)...(((((.((...........)).)))))...(((((....))))))))))).)))..... ( -27.90) >pfo.2296 4419449 120 + 6438405 GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUU ((((((......)))))).........(((.((((((..(..((((.....))))..)...(((((.((...........)).)))))...(((((....))))))))))).)))..... ( -27.90) >consensus GGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGCUGGCUGGGUUCCCCCAUU ((((((......))))))......((((((.(((..((....)))))..))))))......(((((.((...........)).)))))...(((((....))))).((((....)))).. (-29.66 = -28.33 + -1.33)

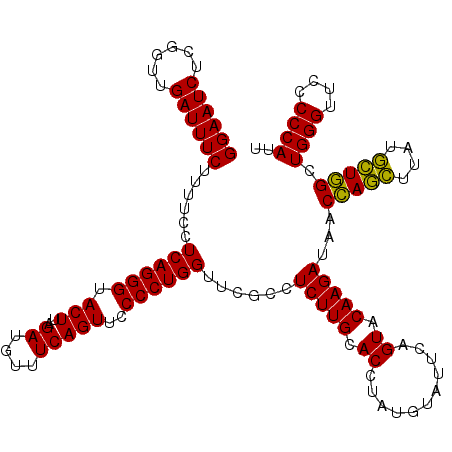
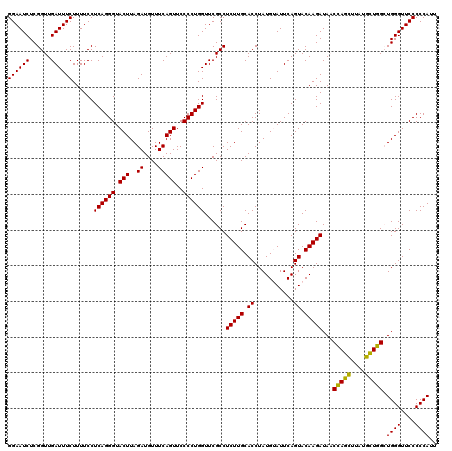
| Location | 5,267,016 – 5,267,136 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -38.67 |
| Energy contribution | -36.90 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.09 |
| Structure conservation index | 1.05 |
| SVM decision value | 6.55 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267016 120 - 6264404 AAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -37.50) >pau.2917 5538948 120 - 6537648 AAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -37.50) >ppk.656 4854312 120 + 6181863 AAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -37.60) >pel.714 4510980 120 + 5888780 AAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -37.60) >pf5.2519 4812886 120 - 7074893 AAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -35.60) >pfo.2296 4419449 120 - 6438405 AAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... ( -35.60) >consensus AAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCC ..(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..)))....... (-38.67 = -36.90 + -1.77)
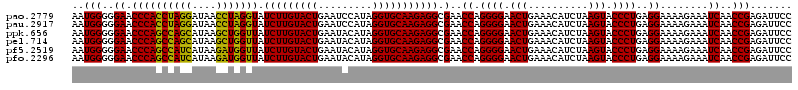
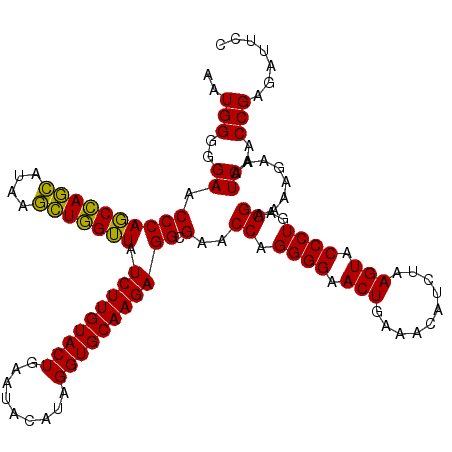
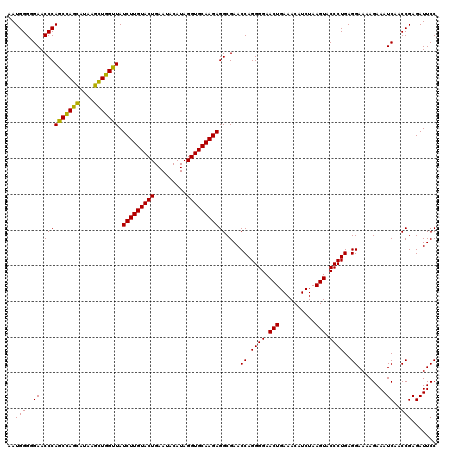
| Location | 5,267,056 – 5,267,176 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -42.65 |
| Energy contribution | -40.60 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.91 |
| Structure conservation index | 1.03 |
| SVM decision value | 6.15 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267056 120 - 6264404 UCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA ((((((((((........)))))...)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ ( -42.90) >pau.2917 5538988 120 - 6537648 UCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA ((((((((((........)))))...)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ ( -42.90) >ppk.656 4854352 120 + 6181863 UUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA (((((.((((........))))....)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ ( -41.10) >pel.714 4511020 120 + 5888780 UUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA (((((.((((........))))....)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ ( -41.10) >pf5.2519 4812926 120 - 7074893 UCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA ((((((((((........)))))...)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ ( -41.00) >pfo.2296 4419489 120 - 6438405 UCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA ((((((((((........))))))..(((((...((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))..)).)))...)))).. ( -40.50) >consensus UCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAA (((((.((((........))))....)))))...((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))........ (-42.65 = -40.60 + -2.05)
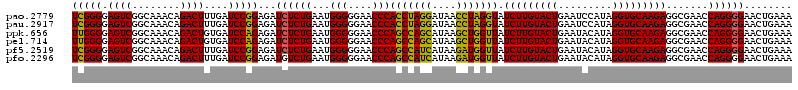

| Location | 5,267,096 – 5,267,216 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -41.53 |
| Consensus MFE | -43.05 |
| Energy contribution | -41.00 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.15 |
| Structure conservation index | 1.04 |
| SVM decision value | 5.17 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267096 120 - 6264404 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUAC ..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))......... ( -43.30) >pau.2917 5539028 120 - 6537648 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUAC ..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))......... ( -43.30) >ppk.656 4854392 120 + 6181863 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUAC ..(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))......... ( -41.50) >pel.714 4511060 120 + 5888780 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUAC ..(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))......... ( -41.50) >pf5.2519 4812966 120 - 7074893 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUAC ..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))......... ( -41.40) >pfo.2296 4419529 120 - 6438405 AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUAC ..(((((.(..........((..(....)..)).....((((((((((((........)))))...))))))).).)))))..(((....)))(((((((....)))))))......... ( -38.20) >consensus AGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUAC ..(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))......... (-43.05 = -41.00 + -2.05)

| Location | 5,267,176 – 5,267,296 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.22 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267176 120 + 6264404 AGCUUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... ( -25.20) >pau.2917 5539108 120 + 6537648 AGCUUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... ( -25.20) >ppk.656 4854472 120 - 6181863 AGCUUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... ( -25.40) >pel.714 4511140 120 - 5888780 AGCUUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... ( -25.40) >pf5.2519 4813046 120 + 7074893 AGCUUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUAUGAAGACGACAUUC (((((.....)))))....((((((.(((.((((.((.....)))))).......(((((.(((......((((............))))......)))))))).)))))))))...... ( -23.20) >pfo.2296 4419609 120 + 6438405 AGCUUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... ( -25.20) >consensus AGCUUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUC (((((.....)))))....((((((.....((((.((.....))))))...(((.(((((.(((......((((............))))......)))))))).)))))))))...... (-24.65 = -24.82 + 0.17)

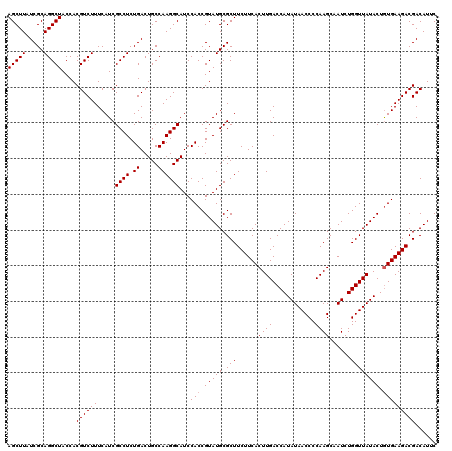
| Location | 5,267,296 – 5,267,398 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -11.16 |
| Energy contribution | -10.38 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267296 102 + 6264404 GCCGAAAAUUCGCG----------CU------UGAACUCGCAAAUUUUACCUUGACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCUUAC--UUCUAUCACAUACCCGAAUUUUUA ...(((((((((((----------..------......)))............((((.((((((((.(........).))))))))))))....--...............)))))))). ( -22.60) >pau.2917 5539228 102 + 6537648 GCCGAAAAUUCGCG----------CU------UGAACUCGCAAAUUUUACCUUGACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCUUAC--UUCUAUCACAUACCCGAAUUUUUA ...(((((((((((----------..------......)))............((((.((((((((.(........).))))))))))))....--...............)))))))). ( -22.60) >ppk.656 4854592 114 - 6181863 GCCGAAAAUUCGUACGUUGCUCUUUC--GAGCAGAACUCACAAAUUUUACCUUAGCCUGAUCCACCAGCAGUGAAACUGG-UGUUCAGUC-UAU--AUCUAUCACAUAUCCGAAUUUUUA ...(((((((((....((((((....--))))))..................(((.((((..((((((........))))-)).)))).)-)).--..............))))))))). ( -29.20) >pel.714 4511260 116 - 5888780 GCCGAAAAUUCGCAUUUCGCUCAAUCAAGAGCAGAACUCACAAAUUUUACCUUAGCCUGAUAAACCACCAGUGAAAGUGG-CCAUCAGUC-UAU--UUCUAUCACAUAUCCGAAUUUUUA ...(((((((((...(((((((......)))).)))................(((.(((((...((((........))))-..))))).)-)).--..............))))))))). ( -24.50) >pf5.2519 4813166 114 + 7074893 GCCGAAAAUUUGCA---UUCUUAAUUAAAAG-AGAACUCACAAAUUUUACCUUAGCCUGAUCCGUUACCAGUGAAAGUAA-CGUUCAGUC-UAUCUUUCUAUCACAUACCCAAAUUUUUA ...(((((((((..---(((((........)-))))................(((.((((..((((((........))))-)).)))).)-)).................))))))))). ( -19.30) >pfo.2296 4419729 112 + 6438405 GCCGAAAAUUCGAA--UUUCUCAACU--AAG-AGAACUCACAAAUUUUACCUUAGCCUGAU-CACCACCAGUGAAAGUGG-CCAUCAGUC-UAUCUUUCUAUCACAUACCCAAAUUUUUA ...(((((((.((.--.(((((....--..)-)))).)).............(((.(((((-..((((........))))-..))))).)-))...................))))))). ( -18.50) >consensus GCCGAAAAUUCGCA___U_CUCAACU___AG_AGAACUCACAAAUUUUACCUUAGCCUGAUCCACCACCAGUGAAAGUGA_CAUUCAGUC_UAU__UUCUAUCACAUACCCGAAUUUUUA ...(((((((((.....................(....).................((((((..((((........))))..))))))......................))))))))). (-11.16 = -10.38 + -0.77)
| Location | 5,267,296 – 5,267,398 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -13.86 |
| Energy contribution | -12.45 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267296 102 - 6264404 UAAAAAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUCA------AG----------CGCGAAUUUUCGGC ..((((((((.((...(((.....--....((((((((((((.((......)).)))))))).))))...(((.....)))....)))------.)----------).)))))))).... ( -27.80) >pau.2917 5539228 102 - 6537648 UAAAAAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUCA------AG----------CGCGAAUUUUCGGC ..((((((((.((...(((.....--....((((((((((((.((......)).)))))))).))))...(((.....)))....)))------.)----------).)))))))).... ( -27.80) >ppk.656 4854592 114 + 6181863 UAAAAAUUCGGAUAUGUGAUAGAU--AUA-GACUGAACA-CCAGUUUCACUGCUGGUGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUGCUC--GAAAGAGCAACGUACGAAUUUUCGGC ..((((((((..(((((.......--.((-(.((((.((-(((((......)))))))..)))).)))...................(((((--....)))))))))))))))))).... ( -36.40) >pel.714 4511260 116 + 5888780 UAAAAAUUCGGAUAUGUGAUAGAA--AUA-GACUGAUGG-CCACUUUCACUGGUGGUUUAUCAGGCUAAGGUAAAAUUUGUGAGUUCUGCUCUUGAUUGAGCGAAAUGCGAAUUUUCGGC .......(((((............--.((-(.(((((((-(((((......))))).))))))).)))......(((((((...(((.((((......)))))))..)))))))))))). ( -31.00) >pf5.2519 4813166 114 - 7074893 UAAAAAUUUGGGUAUGUGAUAGAAAGAUA-GACUGAACG-UUACUUUCACUGGUAACGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCU-CUUUUAAUUAAGAA---UGCAAAUUUUCGGC ..((((((((.((...((((.((((((((-(.((((.((-(((((......)))))))..)))).)))......((((....)))).)-))))).))))...)---).)))))))).... ( -25.20) >pfo.2296 4419729 112 - 6438405 UAAAAAUUUGGGUAUGUGAUAGAAAGAUA-GACUGAUGG-CCACUUUCACUGGUGGUG-AUCAGGCUAAGGUAAAAUUUGUGAGUUCU-CUU--AGUUGAGAAA--UUCGAAUUUUCGGC ..(((((((((((..............((-(.(((((.(-(((((......)))))).-))))).)))................((((-(..--....))))))--)))))))))).... ( -30.80) >consensus UAAAAAUUCGGGUAUGUGAUAGAA__AUA_GACUGAACG_CCACUUUCACUGGUGAUGAAUCAGGCUAAGGUAAAAUUUGUGAGUUCU_CU___AAUUGAG_A___UGCGAAUUUUCGGC ................................((((((..(((((......)))))..))))))(((...(.((((((((((.......(((......))).....)))))))))))))) (-13.86 = -12.45 + -1.41)


| Location | 5,267,320 – 5,267,437 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -11.74 |
| Energy contribution | -10.47 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267320 117 + 6264404 CAAAUUUUACCUUGACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCUUAC--UUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA- .............((((.((((((((.(........).))))))))))))....--..........................(((((.(((((((......))))))).....))))).- ( -28.20) >pau.2917 5539252 117 + 6537648 CAAAUUUUACCUUGACUUGAAUGAUCACCAGUGAAAGAGAUCAUUCAGUCUUAC--UUCUAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCA- .............((((.((((((((.(........).))))))))))))....--..........................(((((.(((((((......))))))).....))))).- ( -28.20) >ppk.656 4854630 112 - 6181863 CAAAUUUUACCUUAGCCUGAUCCACCAGCAGUGAAACUGG-UGUUCAGUC-UAU--AUCUAUCACAUAUCCGAAUUUUUAAAGAAC-GAUCUGAC--AAAAGCCAGAAAUCAACAUUCG- ............(((.((((..((((((........))))-)).)))).)-)).--..............................-..((((..--......))))............- ( -17.80) >pel.714 4511300 112 - 5888780 CAAAUUUUACCUUAGCCUGAUAAACCACCAGUGAAAGUGG-CCAUCAGUC-UAU--UUCUAUCACAUAUCCGAAUUUUUAAAGAAC-GAUCUGAC--AAAAGUCAGAAAUCAACAUUCA- ............(((.(((((...((((........))))-..))))).)-)).--..............................-..((((((--....))))))............- ( -18.70) >pf5.2519 4813202 116 + 7074893 CAAAUUUUACCUUAGCCUGAUCCGUUACCAGUGAAAGUAA-CGUUCAGUC-UAUCUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAUCAAAAGACUAGAAAUCAACAUUCA- ............(((.((((..((((((........))))-)).)))).)-))((.((((.....................)))).-))((((.((....)).))))............- ( -15.10) >pfo.2296 4419764 115 + 6438405 CAAAUUUUACCUUAGCCUGAU-CACCACCAGUGAAAGUGG-CCAUCAGUC-UAUCUUUCUAUCACAUACCCAAAUUUUUAAAGAAC-GAACUAGUC-AAAGACUAGAAAUCAACAUUCAC ............(((.(((((-..((((........))))-..))))).)-))((.((((.....................)))).-)).((((((-...)))))).............. ( -18.60) >consensus CAAAUUUUACCUUAGCCUGAUCCACCACCAGUGAAAGUGA_CAUUCAGUC_UAU__UUCUAUCACAUACCCGAAUUUUUAAAGAAC_GAUCUGAU__AAAGACCAGAAAUCAACAUUCA_ ................((((((..((((........))))..)))))).........................................((((((......))))))............. (-11.74 = -10.47 + -1.27)
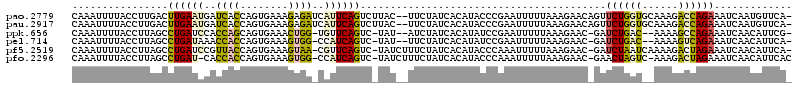
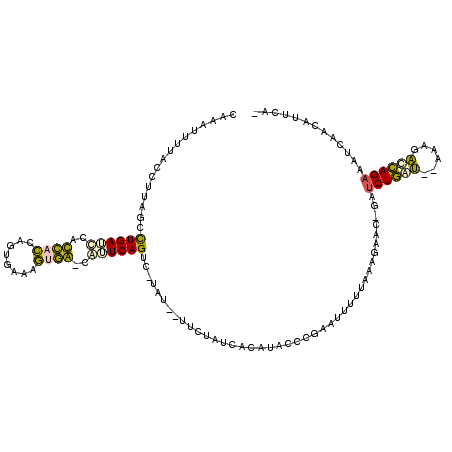
| Location | 5,267,355 – 5,267,463 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -17.14 |
| Energy contribution | -15.34 |
| Covariance contribution | -1.80 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5267355 108 - 6264404 AAAGCUCAGAAAUGAAUGUUCGUGAA----------UGAACAUUGAUUUCUGGUCUUUGCACCAGAACUGUUCUUUAAAAAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCU ((((..(((.(((.((((((((....----------)))))))).)))((((((......)))))).)))..))))....((((((.(((.........--)))...))))))....... ( -24.60) >pau.2917 5539287 108 - 6537648 AAAGCUCAGAAAUGAAUGUUCGUGAA----------UGAACAUUGAUUUCUGGUCUUUGCACCAGAACUGUUCUUUAAAAAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCU ((((..(((.(((.((((((((....----------)))))))).)))((((((......)))))).)))..))))....((((((.(((.........--)))...))))))....... ( -24.60) >ppk.656 4854665 102 + 6181863 AGAACCUGGAAAUGAGCAUUCG-CAU----------CGAAUGUUGAUUUCUGGCUUUU--GUCAGAUC-GUUCUUUAAAAAUUCGGAUAUGUGAUAGAU--AUA-GACUGAACA-CCAGU .....((((.(((.((((((((-...----------)))))))).)))((((((....--))))))..-............(((((.(((((.....))--)))-..)))))..-)))). ( -26.10) >pel.714 4511335 103 + 5888780 AGAACUUAGAAAUGAGCAUUCGGUAG----------UGAAUGUUGAUUUCUGACUUUU--GUCAGAUC-GUUCUUUAAAAAUUCGGAUAUGUGAUAGAA--AUA-GACUGAUGG-CCACU ((((.((((((((.((((((((....----------)))))))).)))))))).))))--(((((.((-.(((((((...((......)).))).))))--...-)))))))..-..... ( -24.50) >pf5.2519 4813237 103 - 7074893 ---GCUUAGAAAUGAGCAUUCA-CUU----------UGAAUGUUGAUUUCUAGUCUUUUGAUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUAGAAAGAUA-GACUGAACG-UUACU ---...(((((((.(((((((.-...----------.))))))).)))))))(((((((.((((.((.-(..(...........)..))).)))).))))))).-.........-..... ( -23.80) >pfo.2296 4419798 112 - 6438405 ---GCUUAGAAAUGAGCAUUCC-AUCGCUGCGAUGGUGAAUGUUGAUUUCUAGUCUUU-GACUAGUUC-GUUCUUUAAAAAUUUGGGUAUGUGAUAGAAAGAUA-GACUGAUGG-CCACU ---((((((((((.((((((((-((((...))))))..)))))).)))))))((((((-..(((...(-((.(((.........))).)))...))))))))).-.......))-).... ( -26.70) >consensus A_AGCUUAGAAAUGAGCAUUCG_CAA__________UGAAUGUUGAUUUCUGGUCUUU__ACCAGAUC_GUUCUUUAAAAAUUCGGGUAUGUGAUAGAA__AUA_GACUGAACG_CCACU ((((.((((((((.((((((((..............)))))))).)))))))).))))........................((((.....................))))......... (-17.14 = -15.34 + -1.80)
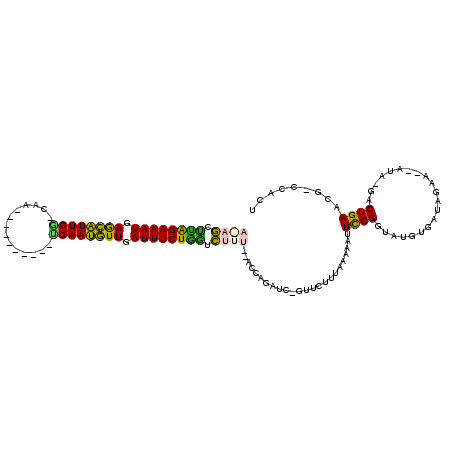
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:15:58 2007