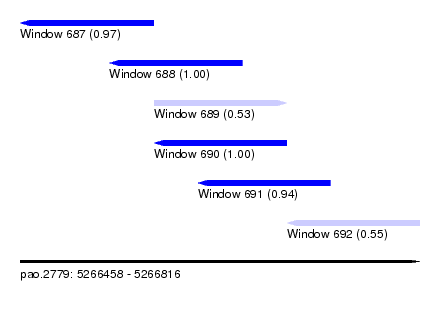
| Sequence ID | pao.2779 |
|---|---|
| Location | 5,266,458 – 5,266,816 |
| Length | 358 |
| Max. P | 0.999872 |

| Location | 5,266,458 – 5,266,578 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.11 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266458 120 - 6264404 UCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG .(((((..(((((.((((...((((((......))))))....)))).....((((((.(((.(.(((...........))).).))).)))))).............)))))..))))) ( -36.90) >pau.2917 5538390 120 - 6537648 UCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG .(((((..(((((.((((...((((((......))))))....)))).....((((((.(((.(.(((...........))).).))).)))))).............)))))..))))) ( -36.90) >ppk.656 4853753 120 + 6181863 UCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG ....((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))..... ( -36.54) >pel.714 4510420 120 + 5888780 UCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG ....((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))..... ( -36.54) >pf5.2519 4812327 120 - 7074893 UCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG ....((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))..... ( -36.54) >pfo.2296 4418890 120 - 6438405 UCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG ....((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))..... ( -36.54) >consensus UCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAG ....((((((((((((((...((((((......))))))....))))...(((.(((.(........).)))))).)).))))))))((.((((..............)))).))..... (-34.66 = -34.11 + -0.55)
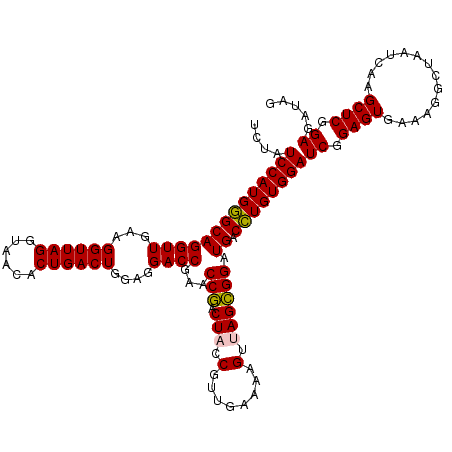
| Location | 5,266,538 – 5,266,657 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -38.49 |
| Energy contribution | -38.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266538 119 - 6264404 UAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUG (((((((.((((((((((.((((((......((((.....)))).-.....)))))).))...(.((((....)))))...)))))).)).)).)))))..((((((......)))))). ( -40.70) >pau.2917 5538470 119 - 6537648 UAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUG (((((((.((((((((((.((((((......((((.....)))).-.....)))))).))...(.((((....)))))...)))))).)).)).)))))..((((((......)))))). ( -40.70) >ppk.656 4853833 119 + 6181863 UAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUG ((((((..((.(((((((...((....))..))))).((((..((-((..((..(.((((....)))))..))..))))..)))))).))..).)))))..((((((......)))))). ( -39.80) >pel.714 4510500 120 + 5888780 UAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUG ...(((....(((...((.(((((((((((..(((....))).)))))...)))))).)).....))).....))).((((((..(....)..))))))..((((((......)))))). ( -40.20) >pf5.2519 4812407 119 - 7074893 UAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUG (((((.....((..((((...((....))..((((.....)))))-)))..))(((.(((.((((((((....))))...)))))))...))).)))))..((((((......)))))). ( -40.20) >pfo.2296 4418970 119 - 6438405 UAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUG (((((.....((..((((...((....))..((((.....)))))-)))..))(((.(((.((((((((....))))...)))))))...))).)))))..((((((......)))))). ( -40.20) >consensus UAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUG (((((........(((((...((....))..))))).....(((......)))(((.((.(((((((((....))))...)))))))...))).)))))..((((((......)))))). (-38.49 = -38.60 + 0.11)

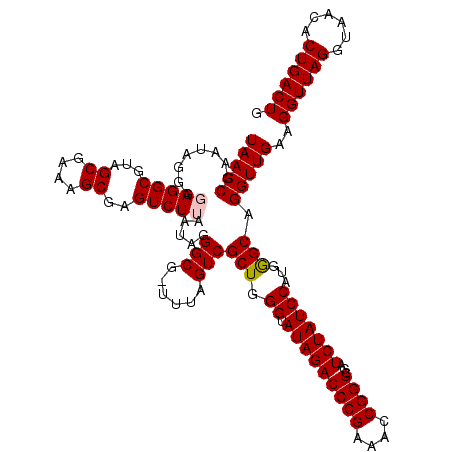
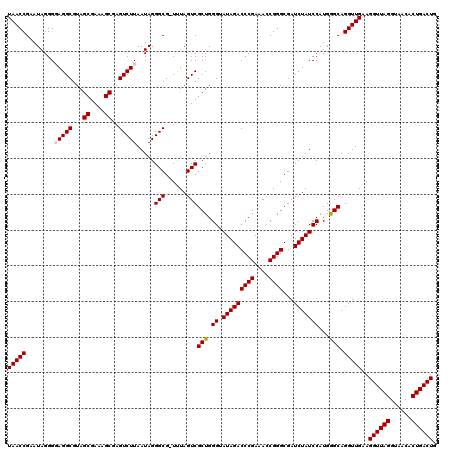
| Location | 5,266,578 – 5,266,697 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266578 119 + 6264404 UCGCCCGGUUUCGGGUCUAUACCCAGCGACUAAA-CGCCCUAUUAAGACUCGCUUUCGCUACGCCUACCCUAUACGGUUAAGCUUGCCACUGAAUAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((...-................((....))...((..(((......)))...))...............)))))))).............. ( -25.30) >pau.2917 5538510 119 + 6537648 UCGCCCGGUUUCGGGUCUAUACCCAGCGACUAAA-CGCCCUAUUAAGACUCGCUUUCGCUACGCCUACCCUAUACGGUUAAGCUUGCCACUGAAUAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((...-................((....))...((..(((......)))...))...............)))))))).............. ( -25.30) >ppk.656 4853873 119 - 6181863 UCGCCCGGUUUCGGGUCUAUACCCAGCGACUAAA-CGCCCUAUUAAGACUCGCUUUCGCUACGCCUCCCCUAUUCGGUUAAGCUCGCCACUGAAUAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((...-................((....))...........((((((((..........))))))))..)))))))).............. ( -26.90) >pel.714 4510540 120 - 5888780 UCGCCCGGUUUCGGGUCUAUACCCAGCGACUAAAGCGCCCUAUUAAGACUCGCUUUCGCUACGCCUCCCCUAUUCGGUUAAGCUCGCCACUGAAUAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((((((((..((....))...))))))..............((((((((..........))))))))..)))))))).............. ( -30.40) >pf5.2519 4812447 119 + 7074893 UCGCCCGGUUUCGGGUCUAUUCCCAGCGACUAGA-CGCCCUAUUAAGACUCGCUUUCGCUACGCCUCCCCUAUUCGGUUAAGCUUGCCACUGAAAAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((.((-.((.((....))....((....))...)).)).....((((((..........))))))....)))))))).............. ( -25.50) >pfo.2296 4419010 119 + 6438405 UCGCCCGGUUUCGGGUCUAUUCCCAGCGACUAGA-CGCCCUAUUAAGACUCGCUUUCGCUACGCCUCCCCUAUUCGGUUAAGCUCGCCACUGAAAAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((.((-.((.((....))....((....))...)).)).....((((((..........))))))....)))))))).............. ( -25.50) >consensus UCGCCCGGUUUCGGGUCUAUACCCAGCGACUAAA_CGCCCUAUUAAGACUCGCUUUCGCUACGCCUCCCCUAUUCGGUUAAGCUCGCCACUGAAUAUAAGUCGCUGACCCAUUAUACAAA ..(((((....))))).......((((((((....................((....))...........((((((((..........))))))))..)))))))).............. (-24.33 = -25.00 + 0.67)

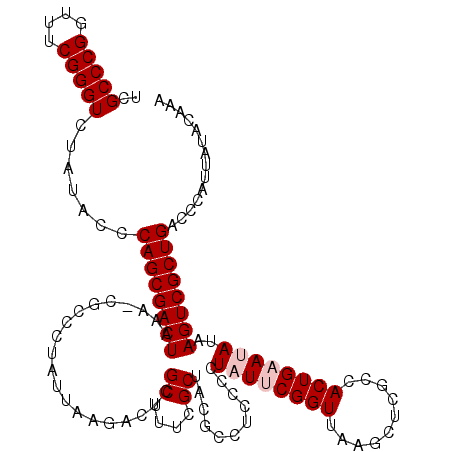
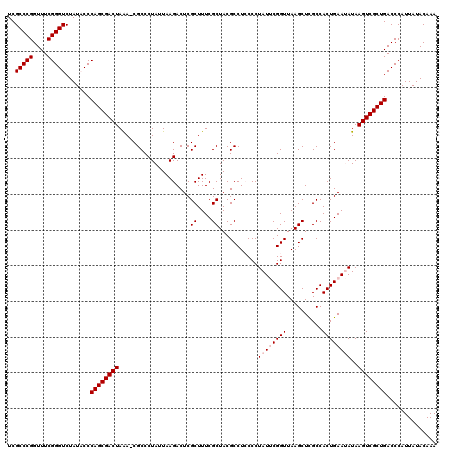
| Location | 5,266,578 – 5,266,697 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -40.22 |
| Energy contribution | -39.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
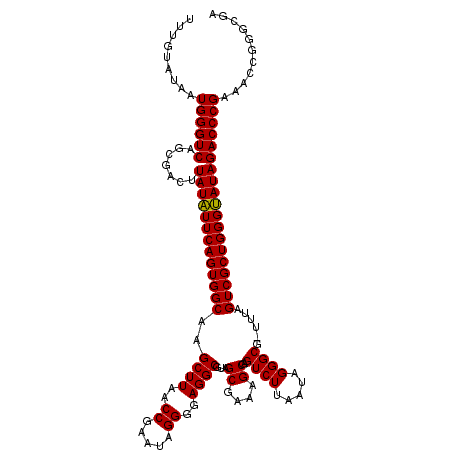
>pao.2779 5266578 119 - 6264404 UUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGA .........((((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))))))))........... ( -40.21) >pau.2917 5538510 119 - 6537648 UUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGA .........((((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))))))))........... ( -40.21) >ppk.656 4853873 119 + 6181863 UUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGA .........((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).-....)))))))))))))))))))........... ( -40.91) >pel.714 4510540 120 + 5888780 UUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGA .........((((((.......(((((((((((((..((((..((.....))..))))((.((....))..((((.....))))))....)))))))))))))))))))........... ( -41.21) >pf5.2519 4812447 119 - 7074893 UUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGA .........((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))))........... ( -40.91) >pfo.2296 4419010 119 - 6438405 UUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGA .........((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))))........... ( -40.91) >consensus UUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGA .........((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....))))......)))))))))))))))))))........... (-40.22 = -39.78 + -0.44)


| Location | 5,266,617 – 5,266,736 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -35.29 |
| Energy contribution | -34.40 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
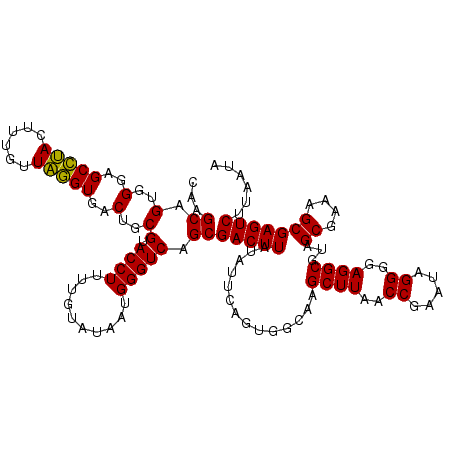
>pao.2779 5266617 119 - 6264404 CAAGCAGUGGGAGCCUACUU-GUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUA ..((((((((....)))).)-)))....((((((.(((((..(((((..(((((..((.(((.......))).))..)))))...)))))))))).))...((....)).))))...... ( -35.40) >pau.2917 5538549 119 - 6537648 CAAGCAGUGGGAGCCUACUU-GUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUA ..((((((((....)))).)-)))....((((((.(((((..(((((..(((((..((.(((.......))).))..)))))...)))))))))).))...((....)).))))...... ( -35.40) >ppk.656 4853912 120 + 6181863 CAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUA ...((.(..(..(((((......)))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....)))))))...... ( -35.40) >pel.714 4510580 120 + 5888780 CAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUA ...((.(..(..(((((......)))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....)))))))...... ( -35.40) >pf5.2519 4812486 120 - 7074893 CAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUA ...((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((.((.....((((..((.....))..))))...)))))))..))))...... ( -35.70) >pfo.2296 4419049 120 - 6438405 CAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUA ...((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))...... ( -36.10) >consensus CAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUA ...((.(..(..(((((......)))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....)))))))...... (-35.29 = -34.40 + -0.89)

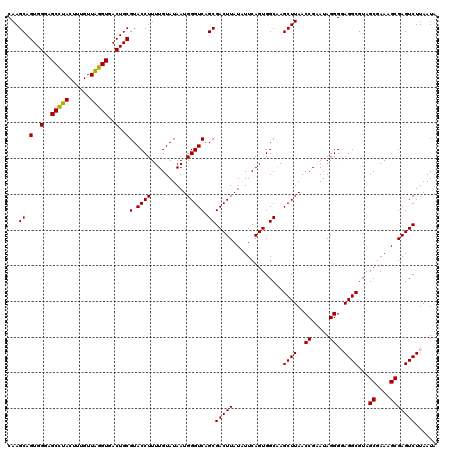
| Location | 5,266,697 – 5,266,816 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -32.52 |
| Energy contribution | -31.63 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.56 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
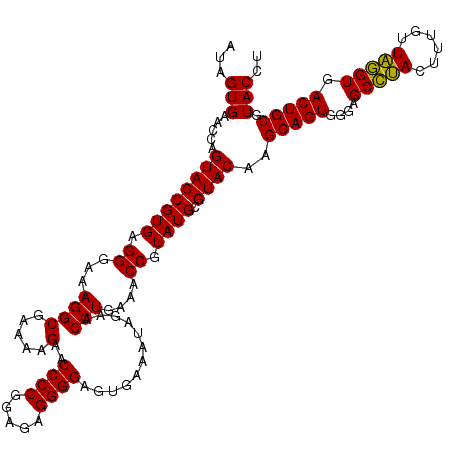
>pao.2779 5266697 119 - 6264404 AUAGUGAACCAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAACCUGAAACCGUAUGCGUACAAGCAGUGGGAGCCUACUU-GUUAGGUGACUGCGUACCU .((((..(((.((((((((.((...((((.....)..((((.....))))............)))....)).)))).)))).((((((((....)))).)-))).))).))))....... ( -32.30) >pau.2917 5538629 119 - 6537648 AUAGUGAACCAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAACCUGAAACCGUAUGCGUACAAGCAGUGGGAGCCUACUU-GUUAGGUGACUGCGUACCU .((((..(((.((((((((.((...((((.....)..((((.....))))............)))....)).)))).)))).((((((((....)))).)-))).))).))))....... ( -32.30) >ppk.656 4853992 120 + 6181863 AUAGUGAACCAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAACCUGAAACCGUAUGCGUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCU ...(((.....((((((((.((...((((.....)..((((.....))))............)))....)).)))).))))..(((((....(((((......))))).))))).))).. ( -31.80) >pel.714 4510660 120 + 5888780 AUAGUGAACCAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAACCUGAAACCGUAUGCGUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCU ...(((.....((((((((.((...((((.....)..((((.....))))............)))....)).)))).))))..(((((....(((((......))))).))))).))).. ( -31.80) >pf5.2519 4812566 120 - 7074893 AUAGUGAACUAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAUCCUGAAACCGUAUGCGUACAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCU .(((....)))((((((((.((...((((.....)..((((.....))))............)))....)).)))).))))..(((((....(((((......))))).)))))...... ( -33.00) >pfo.2296 4419129 120 - 6438405 AUAGUGAACUAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAUCCUGAAACCGUAUGCGUACAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCU .(((....)))((((((((.((...((((.....)..((((.....))))............)))....)).)))).))))..(((((....(((((......))))).)))))...... ( -33.00) >consensus AUAGUGAACCAGUACCGUGAGGGAAAGGCGAAAAGAACCCCGGAGAGGGGAGUGAAAUAGAACCUGAAACCGUAUGCGUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCU ...(((.....((((((((.((...((((.....)..((((.....))))............)))....)).)))).))))..(((((....(((((......))))).))))).))).. (-32.52 = -31.63 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:15:47 2007