
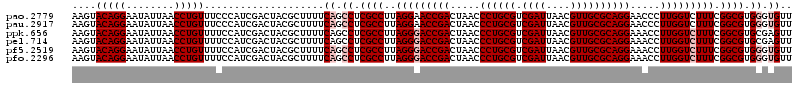
| Sequence ID | pao.2779 |
|---|---|
| Location | 5,265,300 – 5,266,378 |
| Length | 1078 |
| Max. P | 0.999971 |

| Location | 5,265,300 – 5,265,420 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -37.70 |
| Energy contribution | -37.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265300 120 - 6264404 AAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -38.30) >pau.2917 5537232 120 - 6537648 AAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -38.30) >ppk.656 4852594 120 + 6181863 AAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -38.30) >pel.714 4509262 120 + 5888780 AAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -38.30) >pf5.2519 4811169 120 - 7074893 AAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -36.50) >pfo.2296 4417733 120 - 6438405 AAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... ( -36.50) >consensus AAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCUGCA .(((((....((..(((...((....))...........(((..((((((.....))).((((((.......))))))....)))..)))....)))..))(((.....))))))))... (-37.70 = -37.70 + -0.00)

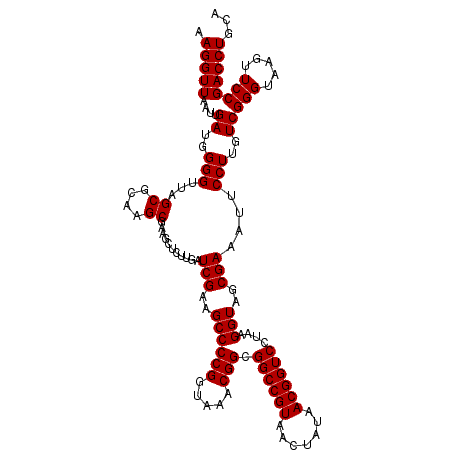
| Location | 5,265,340 – 5,265,460 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -46.70 |
| Energy contribution | -46.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265340 120 - 6264404 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -47.30) >pau.2917 5537272 120 - 6537648 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -47.30) >ppk.656 4852634 120 + 6181863 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -47.30) >pel.714 4509302 120 + 5888780 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -47.30) >pf5.2519 4811209 120 - 7074893 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -45.50) >pfo.2296 4417773 120 - 6438405 GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). ( -45.50) >consensus GAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCU ......((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...)))). (-46.70 = -46.70 + -0.00)


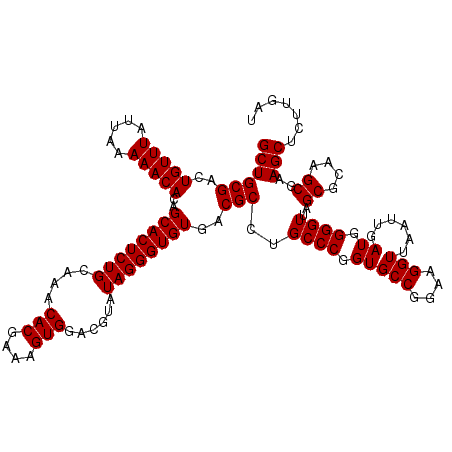
| Location | 5,265,380 – 5,265,500 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -37.30 |
| Energy contribution | -37.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265380 120 - 6264404 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAU (((.((..(((..........((((.(.((.(((...(((....)))...))).))).))))..((..((((.(((((....)))......)).))))..)))))..)).)))....... ( -38.50) >pau.2917 5537312 120 - 6537648 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAU (((.((..(((..........((((.(.((.(((...(((....)))...))).))).))))..((..((((.(((((....)))......)).))))..)))))..)).)))....... ( -38.50) >ppk.656 4852674 120 + 6181863 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAU (((.((..(((..........((((.(.((.(((...(((....)))...))).))).))))..((..((((.(((((....)))......)).))))..)))))..)).)))....... ( -38.50) >pel.714 4509342 120 + 5888780 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAU (((.((..(((..........((((.(.((.(((...(((....)))...))).))).))))..((..((((.(((((....)))......)).))))..)))))..)).)))....... ( -38.50) >pf5.2519 4811249 120 - 7074893 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAU ((((((..(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..))...)))..)))....... ( -37.00) >pfo.2296 4417813 120 - 6438405 GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAU ((((((..(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..))...)))..)))....... ( -37.00) >consensus GCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAU ((((((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..((((.(((((....)))......)).))))..((....))..)))....... (-37.30 = -37.30 + 0.00)

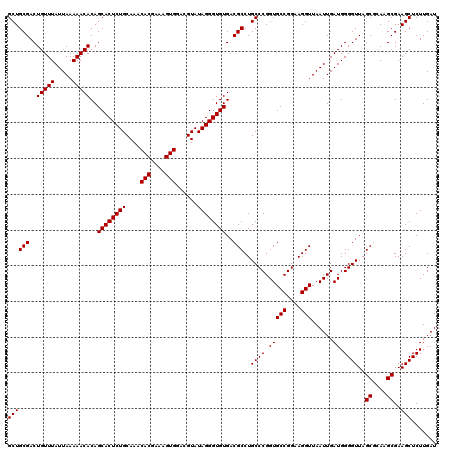
| Location | 5,265,460 – 5,265,580 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -39.19 |
| Energy contribution | -37.47 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.79 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265460 120 - 6264404 ACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..(((((((((((((((((...(((.....)))....))))))))))).))....)))).....((((....(((((.....))))))))).)))......... ( -39.80) >pau.2917 5537392 120 - 6537648 ACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..(((((((((((((((((...(((.....)))....))))))))))).))....)))).....((((....(((((.....))))))))).)))......... ( -39.80) >ppk.656 4852754 120 + 6181863 ACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..((((((((((((.((((...(((.....)))....)))).)))))).))....)))).....((((....(((((.....))))))))).)))......... ( -40.00) >pel.714 4509422 120 + 5888780 ACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..((((((((((((.((((...(((.....)))....)))).)))))).))....)))).....((((....(((((.....))))))))).)))......... ( -39.60) >pf5.2519 4811329 120 - 7074893 ACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..((((((((((((.((((...(((.....)))....)))).)))))).))....)))).....((((....(((((.....))))))))).)))......... ( -34.40) >pfo.2296 4417893 120 - 6438405 ACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..((((((((((((.((((...(((.....)))....)))).)))))).))....)))).....((((....(((((.....))))))))).)))......... ( -39.60) >consensus ACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACAC .(((......)))(((..((((((((((((.((((...(((.....)))....)))).)))))).))....)))).....((((....(((((.....))))))))).)))......... (-39.19 = -37.47 + -1.72)

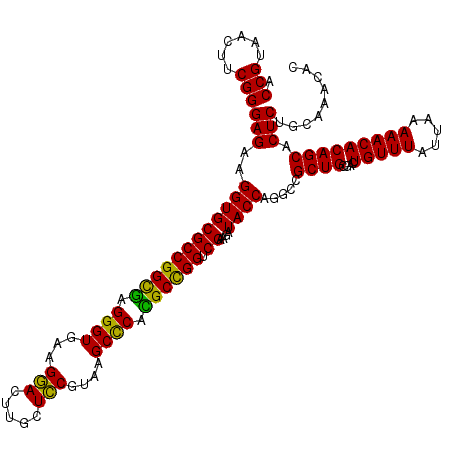
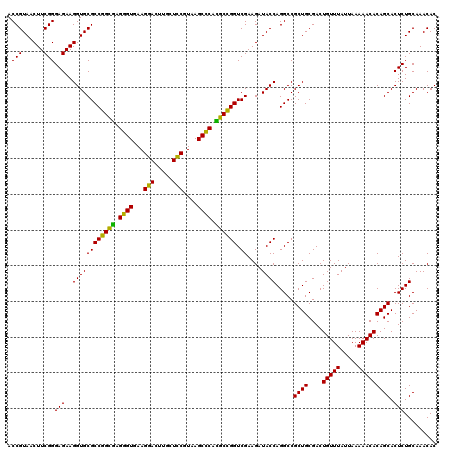
| Location | 5,265,500 – 5,265,620 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -46.57 |
| Consensus MFE | -46.89 |
| Energy contribution | -45.17 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265500 120 - 6264404 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........)).))) ( -47.50) >pau.2917 5537432 120 - 6537648 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........)).))) ( -47.50) >ppk.656 4852794 120 + 6181863 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))) ( -47.70) >pel.714 4509462 120 + 5888780 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))) ( -47.30) >pf5.2519 4811369 120 - 7074893 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))) ( -42.10) >pfo.2296 4417933 120 - 6438405 CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))) ( -47.30) >consensus CGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCACGCCGGUCGAAGAUACCAGGCC ((((((((....)))))))).........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))) (-46.89 = -45.17 + -1.72)

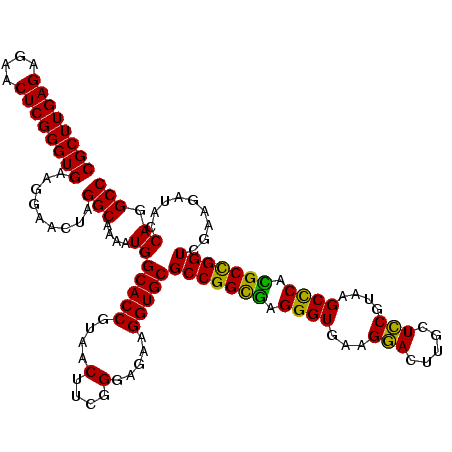
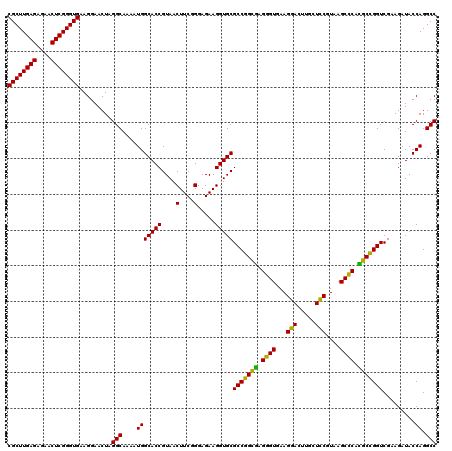
| Location | 5,265,540 – 5,265,660 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -34.07 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265540 120 + 6264404 CCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGG ......(((.(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).((((....).))))))).))). ( -36.70) >pau.2917 5537472 120 + 6537648 CCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGG ......(((.(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).((((....).))))))).))). ( -36.70) >ppk.656 4852834 120 - 6181863 CCUUCACCCUCGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGG (((..(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..))). ( -34.60) >pel.714 4509502 120 - 5888780 GCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGG ......(((..((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))). ( -34.20) >pf5.2519 4811409 120 + 7074893 CCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGG (((..(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..))). ( -34.60) >pfo.2296 4417973 120 + 6438405 GCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGG ......(((..((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))). ( -33.80) >consensus CCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGG ......(((..((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))). (-34.07 = -33.82 + -0.25)

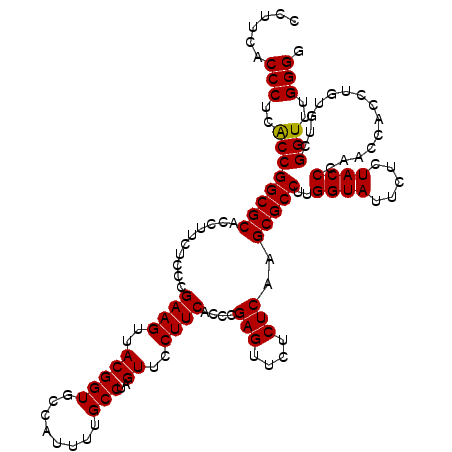
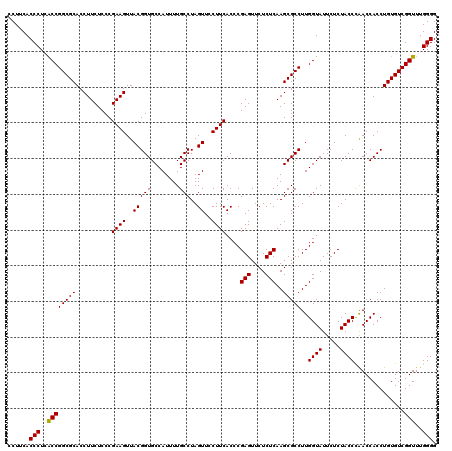
| Location | 5,265,540 – 5,265,660 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -36.07 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265540 120 - 6264404 CCCCAAACCGACACAGGUGGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGG .(((..(((......)))((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).)))))).)))...... ( -39.80) >pau.2917 5537472 120 - 6537648 CCCCAAACCGACACAGGUGGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGG .(((..(((......)))((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).)))))).)))...... ( -39.80) >ppk.656 4852834 120 + 6181863 CCCCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGG .((...(((....((((((.((.((((.....)))).)).))))))......(((((((......))).(((......(((((....(....).....))))))))..)))))))...)) ( -37.90) >pel.714 4509502 120 + 5888780 CCCCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGC .(((..((((.(.((..((.(((((((.....))))....((((((((....)))))))).......))).))...))(((((....(....).....)))))).))))..)))...... ( -39.20) >pf5.2519 4811409 120 - 7074893 CCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGG .(((..(((........(((((.((((.....))))....((((((((....))))))))....)))))(((......(((((....(....).....)))))))))))..)))...... ( -39.00) >pfo.2296 4417973 120 - 6438405 CCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGC .(((..(((........(((((.((((.....))))....((((((((....))))))))....)))))(((......(((((....(....).....)))))))))))..)))...... ( -39.00) >consensus CCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGG .(((...((..((((((((.((.((((.....)))).)).)))))(((....)))..)))..)).....(((......(((((....(....).....)))))))).....)))...... (-36.07 = -36.40 + 0.33)

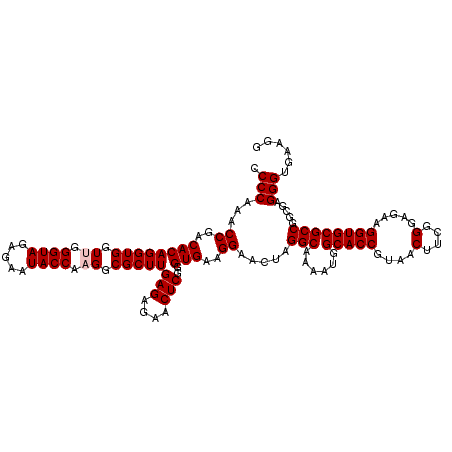
| Location | 5,265,660 – 5,265,780 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -34.10 |
| Energy contribution | -32.27 |
| Covariance contribution | -1.83 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265660 120 - 6264404 CUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAACCAGGAACCGUA ....(((...(((..((((.(.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..))))...).)))).))))))... ( -35.40) >pau.2917 5537592 120 - 6537648 CUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAACCAGGAACCGUA ....(((...(((..((((.(.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..))))...).)))).))))))... ( -35.40) >ppk.656 4852954 119 + 6181863 CUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUCAGGCGACGAAGUGGUUGAUACCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAACUGGGAACCGUA ....((....(((((...((..((((..((((((........((((((-....))))))((((((((....)))..))))).......)))).))..))))..))...)))))..))... ( -33.70) >pel.714 4509622 119 + 5888780 CUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-UUUAGAGCGCGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAACUGGGAACCGUA ....((....(((((.((((..((((..((((((........((((((-....))))))((((((((....)))..))))).......)))).))..)))).))))..)))))..))... ( -35.10) >pf5.2519 4811529 120 - 7074893 CUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACUAGGAACCGUA ....(((...(((.((..(((.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..)))).)))...)).))))))... ( -35.60) >pfo.2296 4418093 119 - 6438405 CUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACCAGGAACCGUA ....(((...(((..((((((.((((..((((((........((((((-....))))))((((((((....)))..))))).......))).)))..))))..)).)))).))))))... ( -37.20) >consensus CUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC_UUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACCAGGAACCGUA ....(((...(((.((..((..((((..((((((........((((((.....))))))((((((((....)))..))))).......)))).))..))))..))...)).))))))... (-34.10 = -32.27 + -1.83)
| Location | 5,265,700 – 5,265,820 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.82 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -41.82 |
| Energy contribution | -39.30 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.15 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.78 |
| SVM RNA-class probability | 0.999949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265700 120 - 6264404 UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUU .(((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..)))))))..... ( -39.60) >pau.2917 5537632 120 - 6537648 UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUU .(((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..)))))))..... ( -39.60) >ppk.656 4852994 119 + 6181863 CUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUCAGGCGACGAAGUGGUUGAUACCAUGCUU ..(((((.(((...(((.(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))..((((((-....))))))....))).....)))))))). ( -39.80) >pel.714 4509662 119 + 5888780 UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-UUUAGAGCGCGAAGUGGUUGAUGCCAUGCUU .(((((((..(..(....(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))..((((((-....))))))...)..)..)))))))..... ( -44.50) >pf5.2519 4811569 120 - 7074893 UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUU .(((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..)))))))..... ( -40.50) >pfo.2296 4418133 119 - 6438405 UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUGCCAUGCUU .(((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((-....))))))...)..)..)))))))..... ( -41.40) >consensus UUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC_UUUAGAUGACGAAGUGGUUGAUGCCAUGCUU .((((((..((((((..(((((....((((....((((((((..((....))..))))))))..)))).)))))...)))))((((((.....)))))).......)..))))))..... (-41.82 = -39.30 + -2.52)
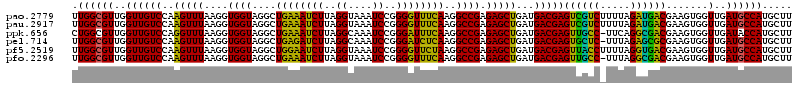
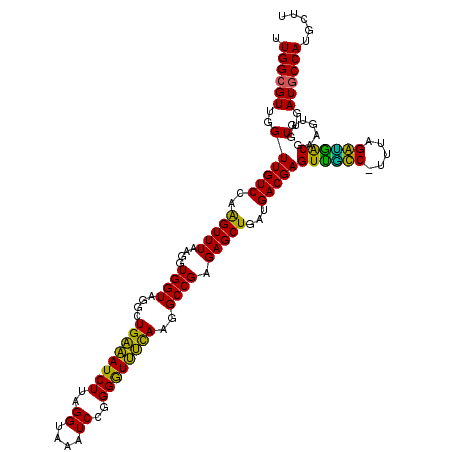
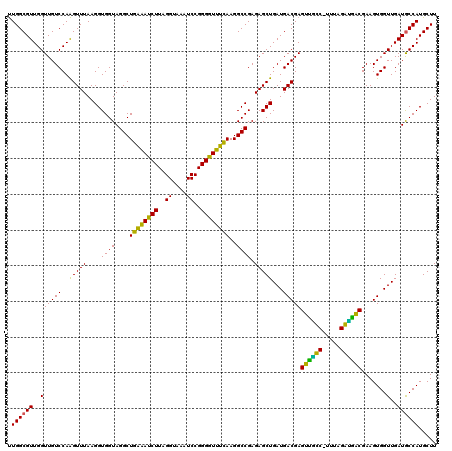
| Location | 5,265,740 – 5,265,860 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -40.70 |
| Energy contribution | -38.82 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265740 120 - 6264404 CUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGAC ..((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))((((((((((((.((.....))))))))))..))))....)))...... ( -38.10) >pau.2917 5537672 120 - 6537648 CUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGAC ..((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))((((((((((((.((.....))))))))))..))))....)))...... ( -38.10) >ppk.656 4853033 120 + 6181863 CCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGAC .(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....))))..... ( -40.30) >pel.714 4509701 120 + 5888780 CCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGAC .(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....))))..... ( -42.80) >pf5.2519 4811609 120 - 7074893 CUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGAC .(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))((((((((((((.((.....))))))))))..))))....))))..... ( -38.50) >pfo.2296 4418172 120 - 6438405 CUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGAC ...((((((((.......((((((..((.((((((....)))))).))..))))))...............((((((((((((.((.....))))))))))..))))....)).)))))) ( -37.60) >consensus CUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGAC .(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....))))..... (-40.70 = -38.82 + -1.88)

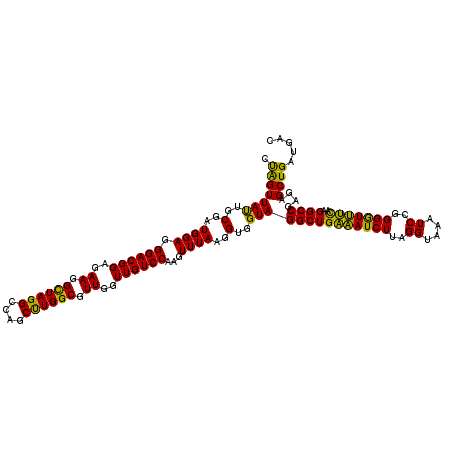
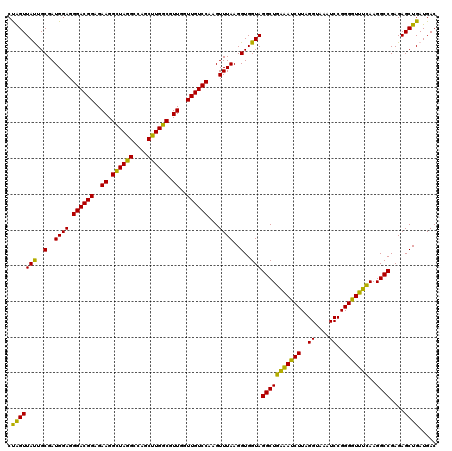
| Location | 5,265,780 – 5,265,900 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -38.18 |
| Energy contribution | -36.85 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.46 |
| SVM RNA-class probability | 0.999903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265780 120 - 6264404 AAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAU ...((((((.(((.(((((((((........)))))..)))).)))))))))......((((((..((.((((((....)))))).))..)))))).((((((......))))))..... ( -39.90) >pau.2917 5537712 120 - 6537648 AAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAU ...((((((.(((.(((((((((........)))))..)))).)))))))))......((((((..((.((((((....)))))).))..)))))).((((((......))))))..... ( -39.90) >ppk.656 4853073 120 + 6181863 AAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAU .....(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))).... ( -38.90) >pel.714 4509741 120 + 5888780 AAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAU .....(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))).... ( -38.70) >pf5.2519 4811649 120 - 7074893 AAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAU .....(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))).... ( -36.40) >pfo.2296 4418212 120 - 6438405 AAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAU .....(((((...((((.(((((........)))))..))))...(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))).... ( -35.20) >consensus AAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAU ...((((((...(((((.(((((........)))))..)))))...))))))......((((((..((.((((((....)))))).))..)))))).((((((......))))))..... (-38.18 = -36.85 + -1.33)

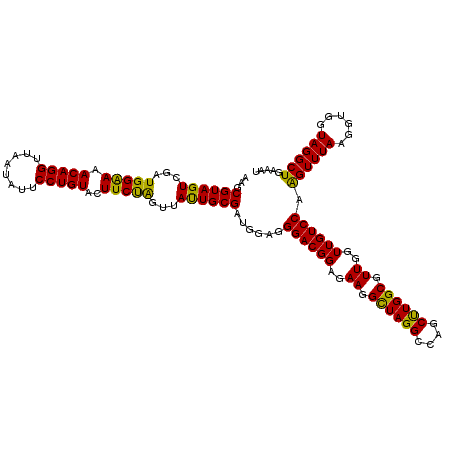
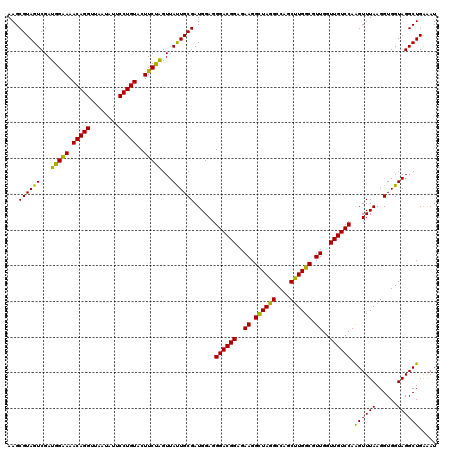
| Location | 5,265,820 – 5,265,940 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -39.53 |
| Consensus MFE | -39.34 |
| Energy contribution | -38.07 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265820 120 - 6264404 AAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....))))))......)))))).)).)) ( -40.10) >pau.2917 5537752 120 - 6537648 AAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....))))))......)))))).)).)) ( -40.10) >ppk.656 4853113 120 + 6181863 AAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).)).....)))).)).)) ( -38.40) >pel.714 4509781 120 + 5888780 AAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)) ( -41.50) >pf5.2519 4811689 120 - 7074893 AAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)) ( -39.20) >pfo.2296 4418252 120 - 6438405 AAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)) ( -37.90) >consensus AAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGC .......((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)) (-39.34 = -38.07 + -1.28)

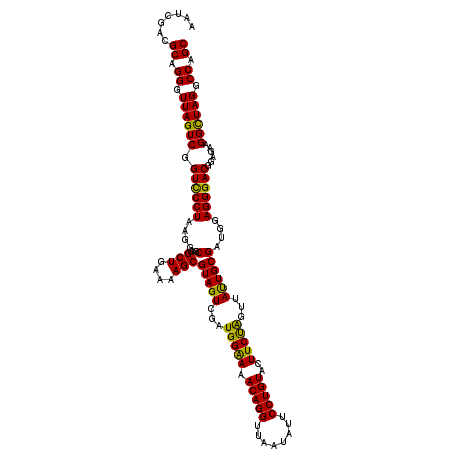
| Location | 5,265,860 – 5,265,980 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -35.40 |
| Energy contribution | -36.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265860 120 + 6264404 AAGUACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUU ....(((((........)))))....................((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).)).. ( -38.10) >pau.2917 5537792 120 + 6537648 AAGUACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUU ....(((((........)))))....................((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).)).. ( -38.10) >ppk.656 4853153 120 - 6181863 AAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUU ....(((((........)))))........((((.(((..........((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).))))))) ( -38.80) >pel.714 4509821 120 - 5888780 AAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUU ....(((((........)))))........((((.(((..........((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).))))))) ( -38.80) >pf5.2519 4811729 120 + 7074893 AAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUU ....(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).)).. ( -40.60) >pfo.2296 4418292 120 + 6438405 AAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUU ....(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).)).. ( -40.60) >consensus AAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUU ....(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).)).. (-35.40 = -36.40 + 1.00)

| Location | 5,265,860 – 5,265,980 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -35.22 |
| Energy contribution | -35.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265860 120 - 6264404 AACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUU ....((((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........))))..(((((........))))).... ( -39.30) >pau.2917 5537792 120 - 6537648 AACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUU ....((((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........))))..(((((........))))).... ( -39.30) >ppk.656 4853153 120 + 6181863 AACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUU ...((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).)))......(((((........))))).... ( -37.00) >pel.714 4509821 120 + 5888780 AACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUU ...((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).)))......(((((........))))).... ( -37.00) >pf5.2519 4811729 120 - 7074893 AACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUU .....(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........))))).... ( -37.60) >pfo.2296 4418292 120 - 6438405 AACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUU .....(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........))))).... ( -37.60) >consensus AACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUU .....(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........))))).... (-35.22 = -35.33 + 0.11)

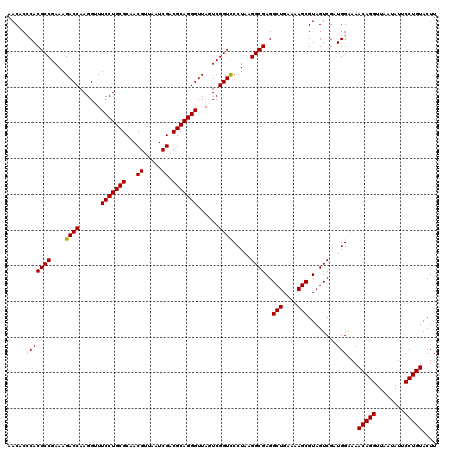
| Location | 5,265,900 – 5,266,020 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -40.35 |
| Energy contribution | -39.80 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265900 120 + 6264404 UUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).....(((((....)).)))....))... ( -39.00) >pau.2917 5537832 120 + 6537648 UUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).....(((((....)).)))....))... ( -39.00) >ppk.656 4853193 120 - 6181863 UUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))... ( -40.90) >pel.714 4509861 120 - 5888780 UUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))... ( -40.90) >pf5.2519 4811769 120 + 7074893 UUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))... ( -41.50) >pfo.2296 4418332 120 + 6438405 UUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))... ( -41.50) >consensus UUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACU ....((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))... (-40.35 = -39.80 + -0.55)

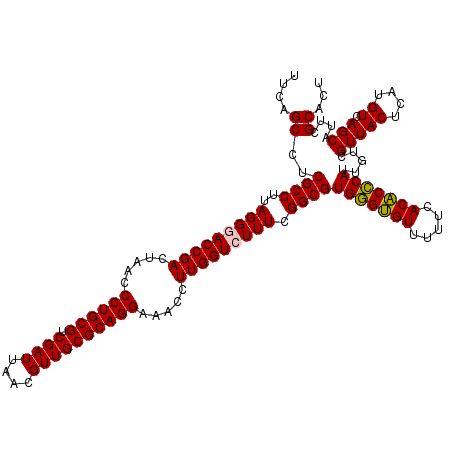
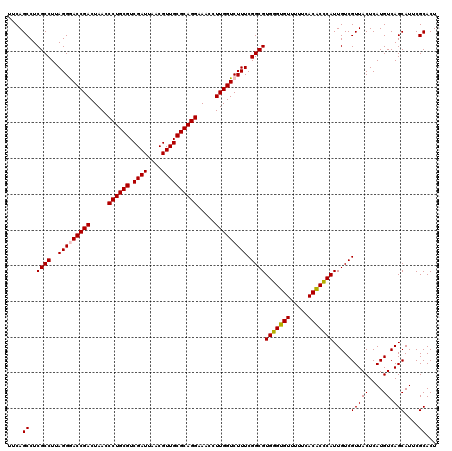
| Location | 5,265,900 – 5,266,020 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -38.91 |
| Energy contribution | -37.80 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265900 120 - 6264404 AGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -36.80) >pau.2917 5537832 120 - 6537648 AGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -36.80) >ppk.656 4853193 120 + 6181863 AGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -38.00) >pel.714 4509861 120 + 5888780 AGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -38.00) >pf5.2519 4811769 120 - 7074893 AGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -38.60) >pfo.2296 4418332 120 - 6438405 AGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ ( -38.60) >consensus AGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAA .((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))........ (-38.91 = -37.80 + -1.11)

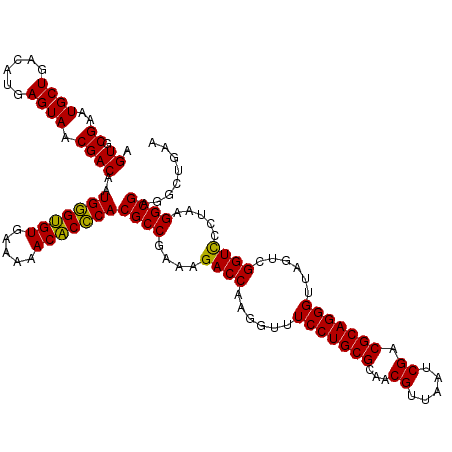
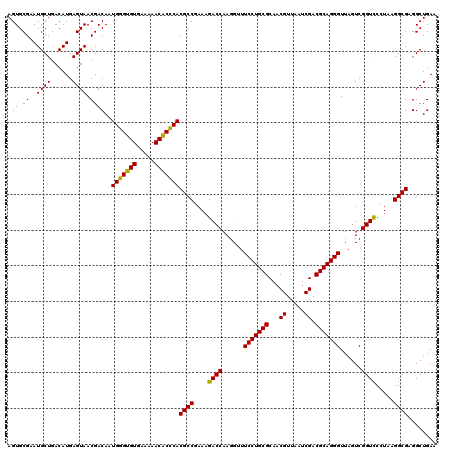
| Location | 5,265,980 – 5,266,098 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -31.89 |
| Energy contribution | -31.17 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265980 118 - 6264404 UGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAA .(((((..--..))))).(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))((((..((.......))..)))).... ( -35.60) >pau.2917 5537912 118 - 6537648 UGUCACGC--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAA .(((((..--..))))).(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))((((..((.......))..)))).... ( -36.60) >ppk.656 4853273 120 + 6181863 UAUCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAA (((((((....)))))))(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).(((..(((.......)))..)))... ( -34.70) >pel.714 4509941 119 + 5888780 UAUCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAA (((((((..-.)))))))(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).(((..(((.......)))..)))... ( -34.60) >pf5.2519 4811849 118 - 7074893 UAUCACGU--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAA ((((((..--..))))))(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))((((..((.......))..)))).... ( -33.80) >pfo.2296 4418412 118 - 6438405 UAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAA ..((((((--...(.((((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).))).)((....)).....)))))).. ( -35.00) >consensus UAUCACGU__AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAA ((((((......))))))(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).(((..((....)).)))......... (-31.89 = -31.17 + -0.72)

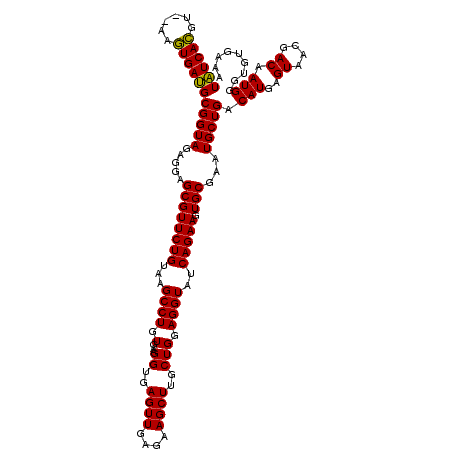

| Location | 5,266,020 – 5,266,138 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -39.45 |
| Energy contribution | -38.73 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266020 118 - 6264404 AAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -41.90) >pau.2917 5537952 118 - 6537648 AAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGC--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -42.90) >ppk.656 4853313 120 + 6181863 AAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -39.30) >pel.714 4509981 119 + 5888780 AAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))((((((((..-.)))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -39.20) >pf5.2519 4811889 118 - 7074893 AAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGU--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -40.20) >pfo.2296 4418452 118 - 6438405 AAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... ( -41.20) >consensus AAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGU__AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGA ..(((...(((((((((..(((.....(((......)))(((((((......))))))))))....))))..)))))..((((.(...((..((((....))))..))).)))))))... (-39.45 = -38.73 + -0.72)


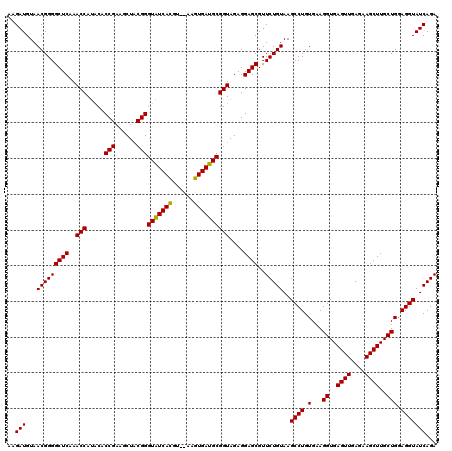
| Location | 5,266,060 – 5,266,178 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -37.55 |
| Energy contribution | -36.83 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266060 118 - 6264404 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))... ( -40.00) >pau.2917 5537992 118 - 6537648 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGC--AAGUGACGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))... ( -41.00) >ppk.656 4853353 120 + 6181863 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))... ( -37.40) >pel.714 4510021 119 + 5888780 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((..-.)))))))))))....))))..)))))... ( -37.30) >pf5.2519 4811929 118 - 7074893 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGU--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))... ( -38.30) >pfo.2296 4418492 118 - 6438405 AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))... ( -39.30) >consensus AGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGU__AAGUGAUGCGGUAGAGGAGCGUUCUGUAAG .....(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((......))))))))))....))))..)))))... (-37.55 = -36.83 + -0.72)
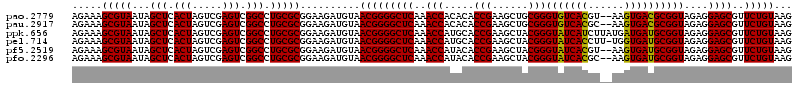
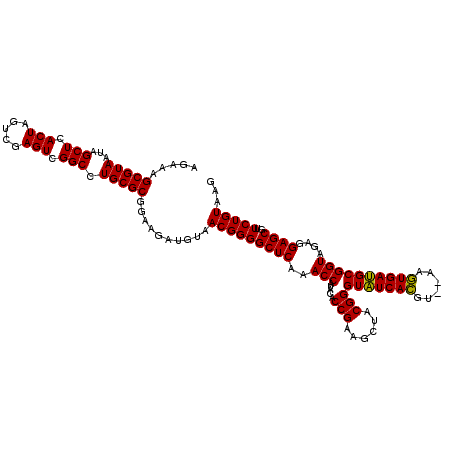
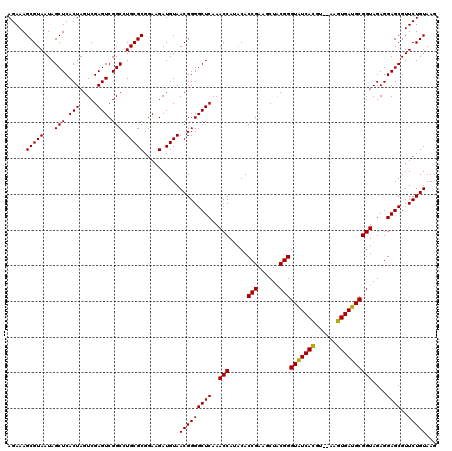
| Location | 5,266,178 – 5,266,298 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266178 120 + 6264404 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..))).))))))....... ( -32.60) >pau.2917 5538110 120 + 6537648 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..))).))))))....... ( -32.60) >ppk.656 4853473 120 - 6181863 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).))))))....... ( -36.60) >pel.714 4510140 120 - 5888780 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).))))))....... ( -36.60) >pf5.2519 4812047 120 + 7074893 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))....... ( -33.64) >pfo.2296 4418610 120 + 6438405 UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))....... ( -33.64) >consensus UUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGAC ..((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))....... (-33.78 = -33.34 + -0.44)

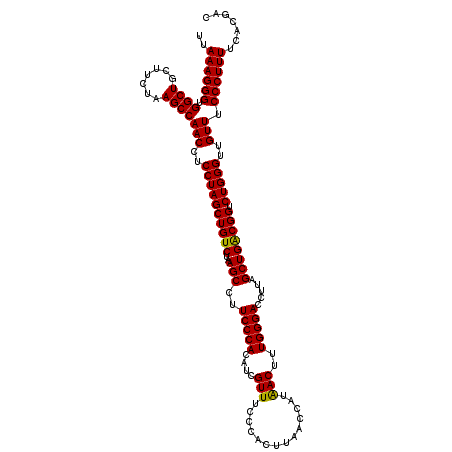
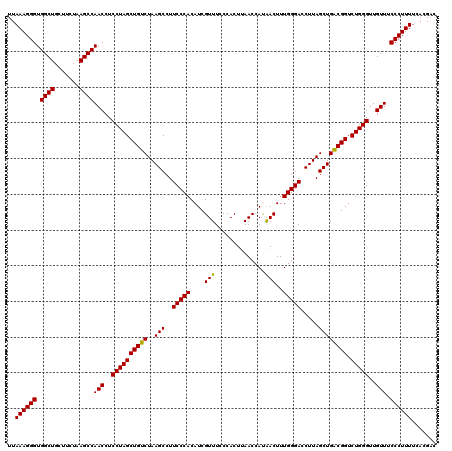
| Location | 5,266,178 – 5,266,298 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -35.84 |
| Energy contribution | -35.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266178 120 - 6264404 GUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((.((((..(....)..))))))))))..((.....))....))))..((((....(((((....)))))(((....))))))).)))))).. ( -36.00) >pau.2917 5538110 120 - 6537648 GUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((.((((..(....)..))))))))))..((.....))....))))..((((....(((((....)))))(((....))))))).)))))).. ( -36.00) >ppk.656 4853473 120 + 6181863 GUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).. ( -35.10) >pel.714 4510140 120 + 5888780 GUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).. ( -35.10) >pf5.2519 4812047 120 - 7074893 GUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).. ( -36.90) >pfo.2296 4418610 120 - 6438405 GUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).. ( -36.90) >consensus GUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAA .......((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).. (-35.84 = -35.73 + -0.11)

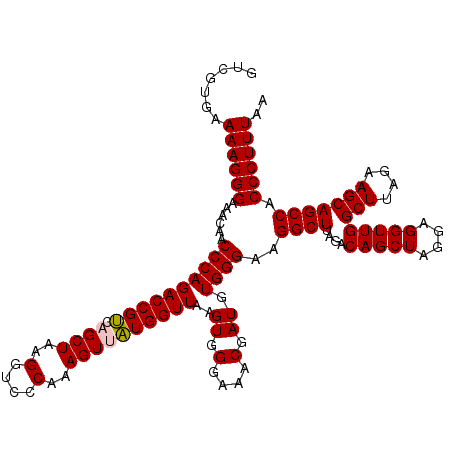
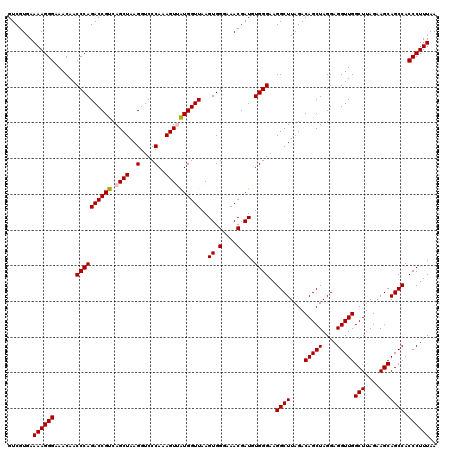
| Location | 5,266,218 – 5,266,338 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -40.91 |
| Consensus MFE | -39.34 |
| Energy contribution | -39.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266218 120 - 6264404 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUU ...(((.(((.(((......((((((((..(....)..))))))))....(((......)))......))))))....(((((..(((((.............))))).))))).))).. ( -41.32) >pau.2917 5538150 120 - 6537648 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUU ...(((.(((.(((......((((((((..(....)..))))))))....(((......)))......))))))....(((((..(((((.............))))).))))).))).. ( -41.32) >ppk.656 4853513 120 + 6181863 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUU ...(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).. ( -41.10) >pel.714 4510180 120 + 5888780 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUU ...(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).. ( -41.10) >pf5.2519 4812087 120 - 7074893 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUU ...(((..((((.(......((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))...))))...).)))).....))).. ( -40.30) >pfo.2296 4418650 120 - 6438405 AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUU ...(((..((((.(......((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))...))))...).)))).....))).. ( -40.30) >consensus AGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUU ...(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).. (-39.34 = -39.23 + -0.11)

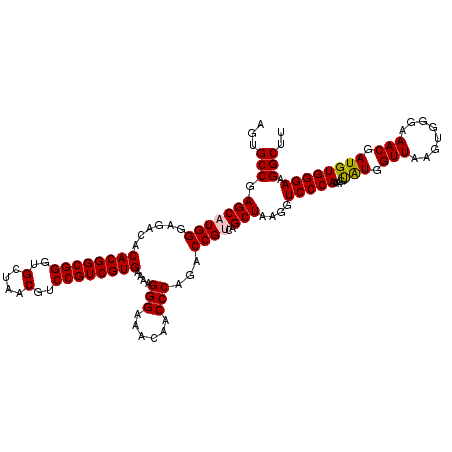
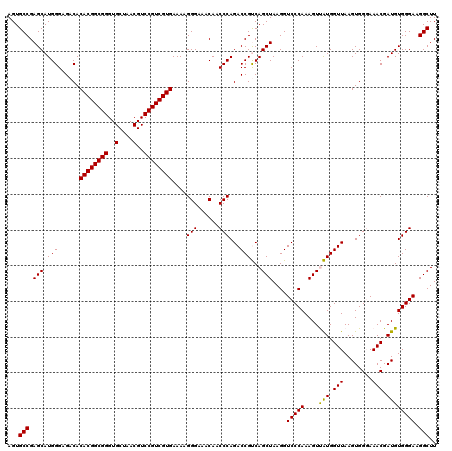
| Location | 5,266,258 – 5,266,378 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5266258 120 - 6264404 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUC ((((...........((.((((..(((.(.........).))).....)))).)).....((((((((..(....)..))))))))....)))).........((((.........)))) ( -34.60) >pau.2917 5538190 120 - 6537648 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUC ((((...........((.((((..(((.(.........).))).....)))).)).....((((((((..(....)..))))))))....)))).........((((.........)))) ( -34.60) >ppk.656 4853553 120 + 6181863 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUC ....(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))) ( -34.42) >pel.714 4510220 120 + 5888780 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUC ....(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))) ( -34.42) >pf5.2519 4812127 120 - 7074893 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUC ....(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))) ( -34.42) >pfo.2296 4418690 120 - 6438405 UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUC ....(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))) ( -34.42) >consensus UCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUC ((((...........((.((((..(((.............))).....)))).)).....((((((((..(....)..))))))))....)))).........((((.........)))) (-33.32 = -33.32 + 0.00)

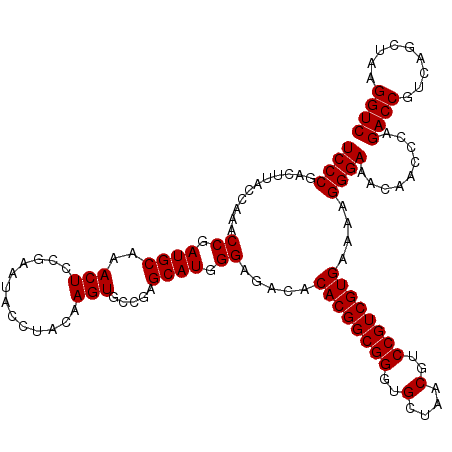
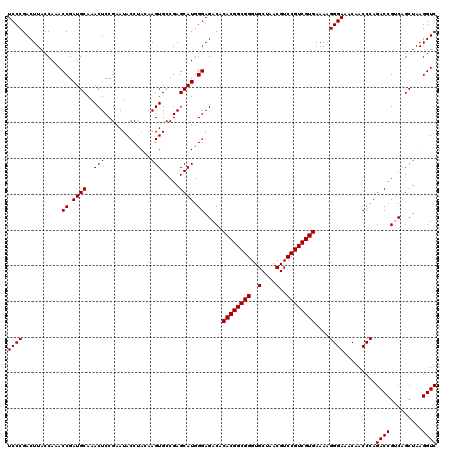
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:15:40 2007