
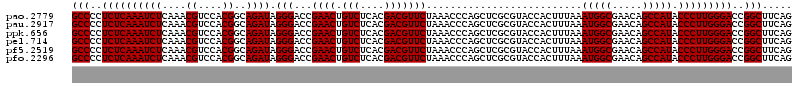
| Sequence ID | pao.2779 |
|---|---|
| Location | 5,264,460 – 5,265,180 |
| Length | 720 |
| Max. P | 0.999911 |

| Location | 5,264,460 – 5,264,580 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -37.07 |
| Energy contribution | -35.68 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
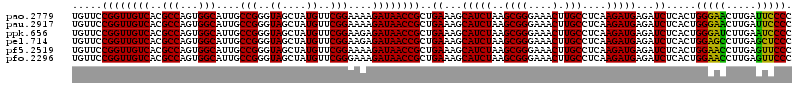
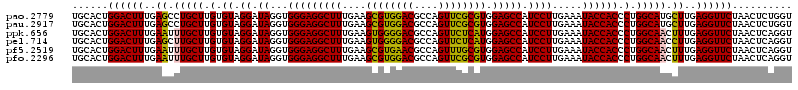
>pao.2779 5264460 120 - 6264404 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAACUUGAUUCCCC .((((((((((((..(((...)))....(((..((....))..)))....)))))))...((...(((((..((((....).)))....)))))...))......))))).......... ( -36.10) >pau.2917 5536392 120 - 6537648 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAACUUGAUUCCCC .((((((((((((..(((...)))....(((..((....))..)))....)))))))...((...(((((..((((....).)))....)))))...))......))))).......... ( -36.10) >ppk.656 4851754 120 + 6181863 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAUCUUGAAUCCCC ...((((((((((((....))))))..))))))((((....(((....)))......))))...((......))((((..((((...))))..(((((((.....)))))))...)))). ( -39.40) >pel.714 4508422 120 + 5888780 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAGCCUUGAGCUCCC ..(((((((((((((....))))))..)))(..((....))..)))))(((((..(((((...........))))).........(((.....))))))))...(((((.....))))). ( -40.30) >pf5.2519 4810329 120 - 7074893 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUUCCC .((((((((.(((....((((((.(((.(((..((....))..)))....)))..))))))....(((((..((((....).)))....))))).)))...))))))))........... ( -36.10) >pfo.2296 4416893 120 - 6438405 UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUUCCC ......(((((((((....))))))..)))(((.((((..((((.((..((((..(((((...........))))).........(((.....)))))))..)).))))....))))))) ( -37.50) >consensus UGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUCCCC .....((((((((..(((...)))....(((..((....))..)))....))))))))..((...(((((..((((....).)))....)))))...)).....(((((.....))))). (-37.07 = -35.68 + -1.39)
| Location | 5,264,500 – 5,264,620 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -42.13 |
| Energy contribution | -42.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
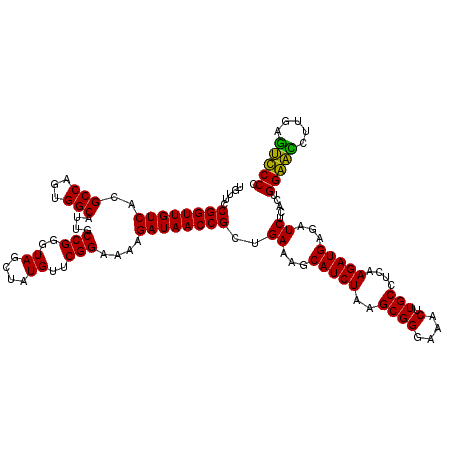
>pao.2779 5264500 120 - 6264404 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA ((((..((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....)))))))))))..))))(((.....))).. ( -42.10) >pau.2917 5536432 120 - 6537648 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA ((((..((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....)))))))))))..))))(((.....))).. ( -42.10) >ppk.656 4851794 120 + 6181863 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA .(((((..(((((.(((.(((((((.(......)))))))).)))))..((((((....)))))).)))..)).)))....(((....)))....(((((...........))))).... ( -44.50) >pel.714 4508462 120 + 5888780 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA .(((((..(((((.(((.(((((((.(......)))))))).)))))..((((((....)))))).)))..)).)))....(((....)))....(((((...........))))).... ( -44.50) >pf5.2519 4810369 120 - 7074893 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA ((((..((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....)))))))))))..))))(((.....))).. ( -42.10) >pfo.2296 4416933 120 - 6438405 UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA ((((..((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...))).((.(((..((....))..))).)).)))))))))))..))))(((.....))).. ( -43.30) >consensus UGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAA ((((..((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....)))))))))))..))))(((.....))).. (-42.13 = -42.13 + -0.00)

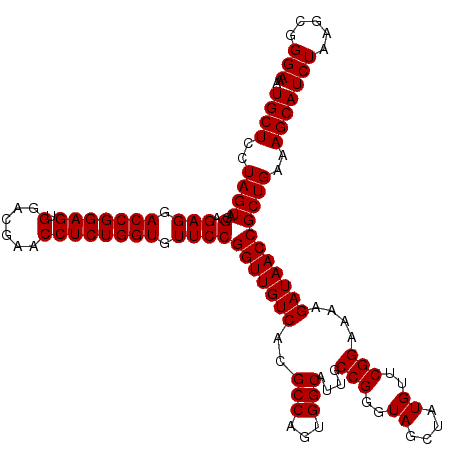
| Location | 5,264,620 – 5,264,740 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.87 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264620 120 + 6264404 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >pau.2917 5536552 120 + 6537648 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >ppk.656 4851914 120 - 6181863 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >pel.714 4508582 120 - 5888780 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >pf5.2519 4810489 120 + 7074893 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >pfo.2296 4417053 120 + 6438405 GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... ( -31.00) >consensus GCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAG (((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))..... (-31.00 = -31.00 + 0.00)
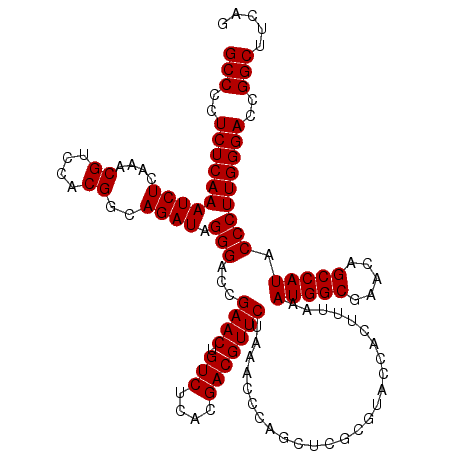
| Location | 5,264,660 – 5,264,780 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -50.30 |
| Consensus MFE | -50.30 |
| Energy contribution | -50.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264660 120 - 6264404 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >pau.2917 5536592 120 - 6537648 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >ppk.656 4851954 120 + 6181863 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >pel.714 4508622 120 + 5888780 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >pf5.2519 4810529 120 - 7074893 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >pfo.2296 4417093 120 - 6438405 GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) ( -50.30) >consensus GGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUC ((((.....))))......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))) (-50.30 = -50.30 + 0.00)

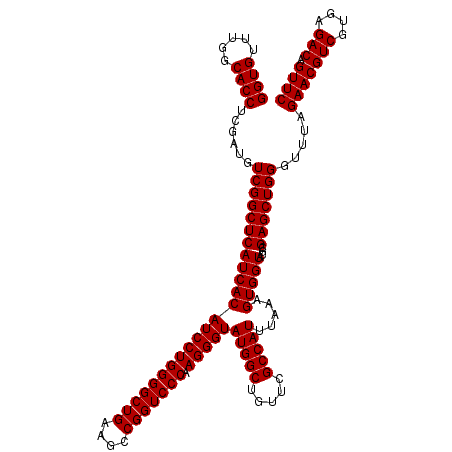
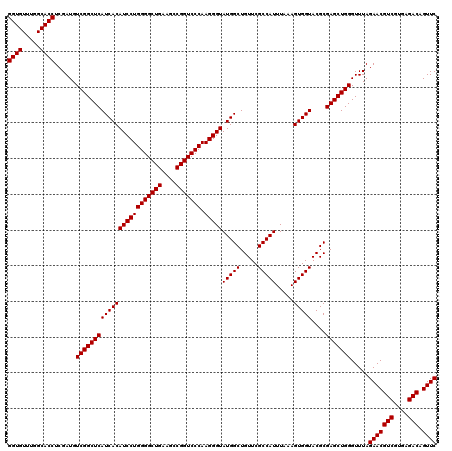
| Location | 5,264,700 – 5,264,820 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -47.20 |
| Energy contribution | -47.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264700 120 - 6264404 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >pau.2917 5536632 120 - 6537648 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >ppk.656 4851994 120 + 6181863 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >pel.714 4508662 120 + 5888780 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >pf5.2519 4810569 120 - 7074893 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >pfo.2296 4417133 120 - 6438405 GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... ( -47.20) >consensus GAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAA .......(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))....... (-47.20 = -47.20 + -0.00)

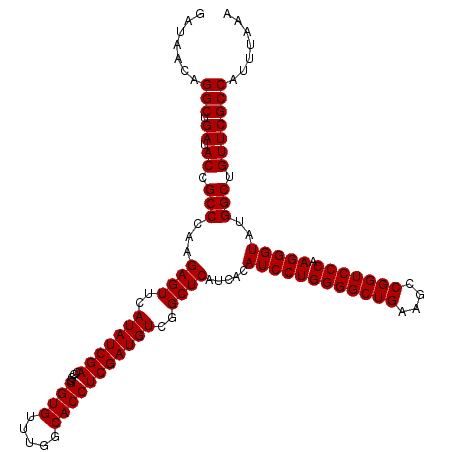
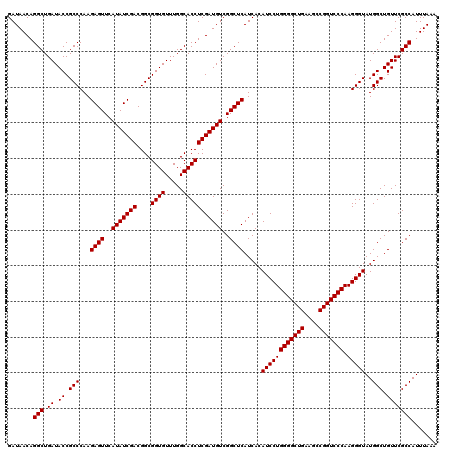
| Location | 5,264,820 – 5,264,940 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.56 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -37.80 |
| Energy contribution | -37.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264820 120 - 6264404 AAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((........((((((((((......).)).)))))))...(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -38.60) >pau.2917 5536752 120 - 6537648 AAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((........((((((((((......).)).)))))))...(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -38.60) >ppk.656 4852114 120 + 6181863 AAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((..............((((..(((......)))..)))).(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -37.80) >pel.714 4508782 120 + 5888780 AAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((..............((((..(((......)))..)))).(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -37.80) >pf5.2519 4810689 120 - 7074893 AAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((........((((((((((......).)).)))))))...(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -38.60) >pfo.2296 4417253 120 - 6438405 AAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((........((((((((((......).)).)))))))...(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. ( -38.60) >consensus AAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGG .(((((..............((((..(((......)))..)))).(((....))).)))))..(((..((((((((((.......)))))))))).))).((((..........)))).. (-37.80 = -37.80 + 0.00)

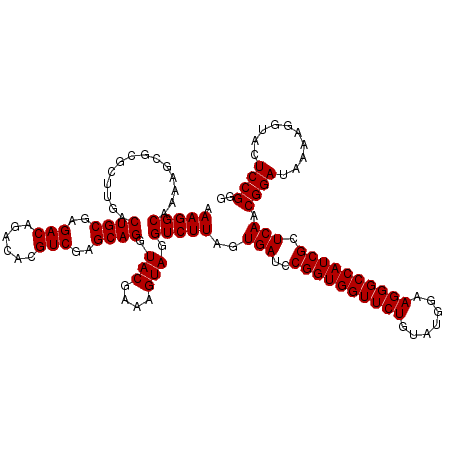
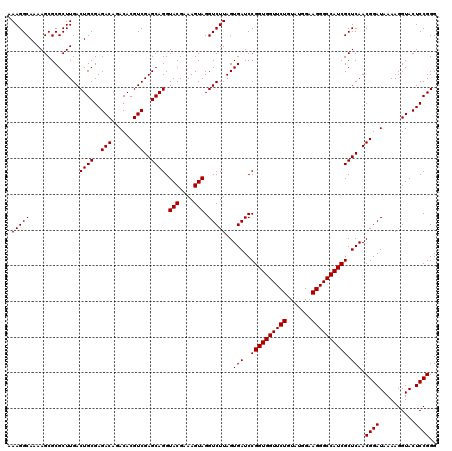
| Location | 5,264,860 – 5,264,980 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -40.40 |
| Energy contribution | -40.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264860 120 - 6264404 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -41.00) >pau.2917 5536792 120 - 6537648 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -40.40) >ppk.656 4852154 120 + 6181863 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -40.40) >pel.714 4508822 120 + 5888780 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -40.40) >pf5.2519 4810729 120 - 7074893 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -40.40) >pfo.2296 4417293 120 - 6438405 AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). ( -40.40) >consensus AAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGU .(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))(((.....))). (-40.40 = -40.40 + -0.00)

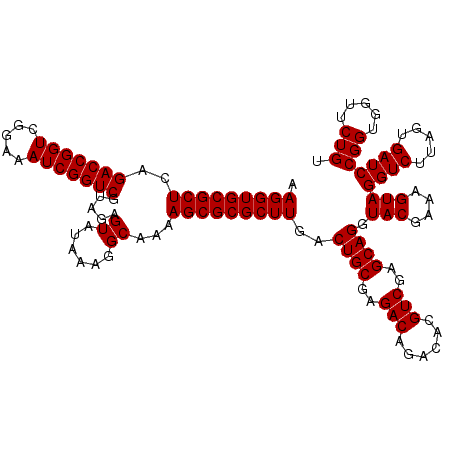
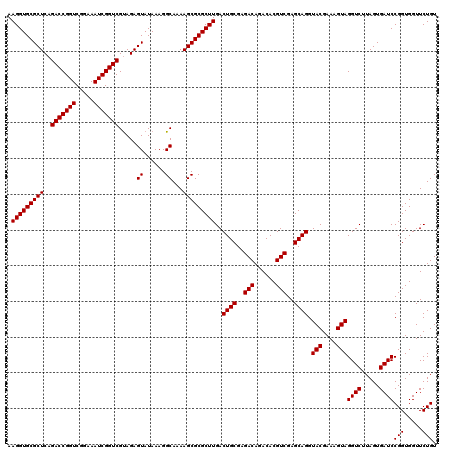
| Location | 5,264,900 – 5,265,020 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -34.00 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264900 120 + 6264404 UCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGU ..((((((.......))).)))..((((((((..((...........))(((((.........))))).))))))......((.(((((((...........))))))))).))...... ( -35.60) >pau.2917 5536832 120 + 6537648 UCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGU ...(((((.(((.....))).)).((((((....)).............(((((.........)))))...)))).....))).(((((((...........)))))))........... ( -34.40) >ppk.656 4852194 120 - 6181863 UCGACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGU ..((((....))))..((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))...... ( -34.10) >pel.714 4508862 120 - 5888780 UCGACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGU ..((((....))))..((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))...... ( -34.10) >pf5.2519 4810769 120 + 7074893 UCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGU ...(((((.(((.....))).)).((((((....)).............(((((.........)))))...)))).....))).(((((((...........)))))))........... ( -34.40) >pfo.2296 4417333 120 + 6438405 UCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGU ...(((((.(((.....))).)).((((((....)).............(((((.........)))))...)))).....))).(((((((...........)))))))........... ( -34.40) >consensus UCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGU ..((((((.......))).)))..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))....... (-34.00 = -34.00 + 0.00)

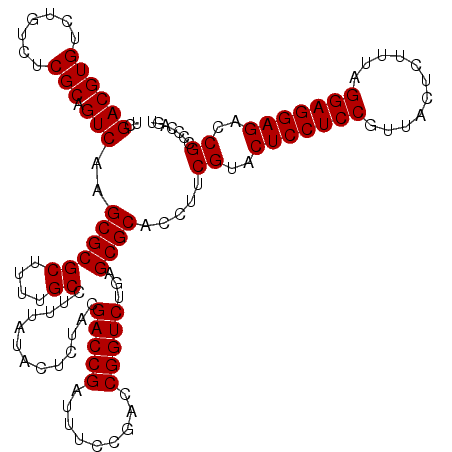
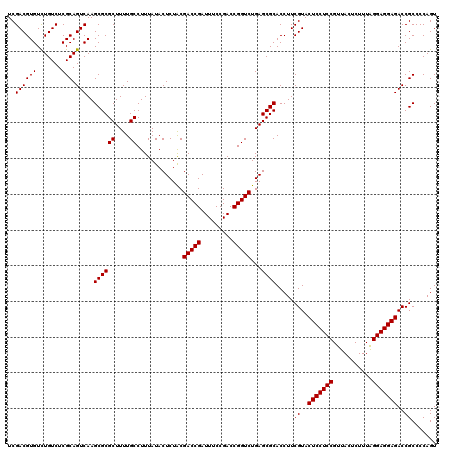
| Location | 5,264,900 – 5,265,020 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -43.93 |
| Energy contribution | -43.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264900 120 - 6264404 ACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....)))))))((...........))...)))))))))))))))..(((......))).. ( -44.70) >pau.2917 5536832 120 - 6537648 ACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. ( -44.10) >ppk.656 4852194 120 + 6181863 ACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. ( -43.60) >pel.714 4508862 120 + 5888780 ACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. ( -43.60) >pf5.2519 4810769 120 - 7074893 ACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. ( -44.10) >pfo.2296 4417333 120 - 6438405 ACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. ( -44.10) >consensus ACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGA ......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).. (-43.93 = -43.93 + -0.00)

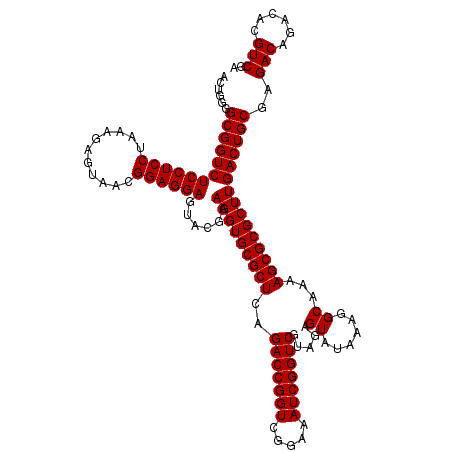
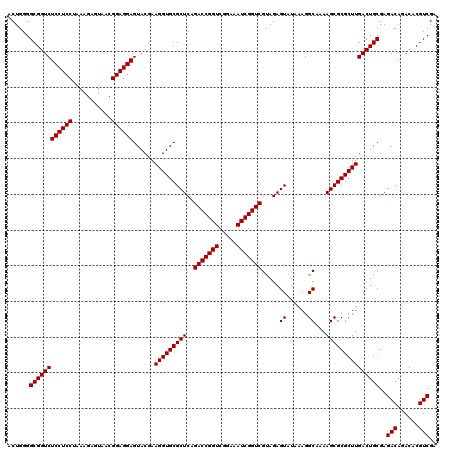
| Location | 5,264,940 – 5,265,060 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264940 120 + 6264404 AUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))...(((.....)))............))).)))))))))))...))) ( -31.00) >pau.2917 5536872 120 + 6537648 AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUCACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))...(((.....)))............))).)))))))))))...))) ( -31.00) >ppk.656 4852234 120 - 6181863 AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))....((.....)).............))).)))))))))))...))) ( -28.10) >pel.714 4508902 120 - 5888780 AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))....((.....)).............))).)))))))))))...))) ( -28.10) >pf5.2519 4810809 120 + 7074893 AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))....((.....)).............))).)))))))))))...))) ( -28.70) >pfo.2296 4417373 120 + 6438405 AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))....((.....)).............))).)))))))))))...))) ( -28.70) >consensus AUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGG ...........(((...((((...((((.(((.(((....((..(((((((...........)))))))..))....((.....)).............))).)))))))))))...))) (-28.50 = -28.50 + -0.00)

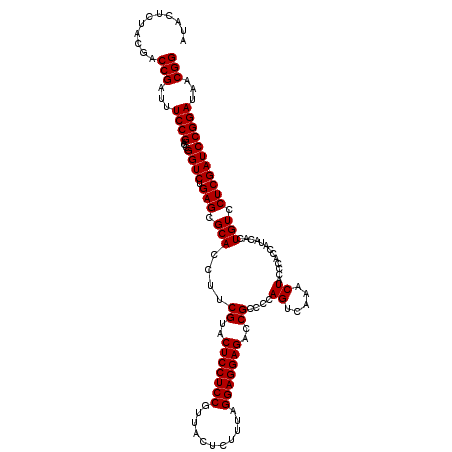
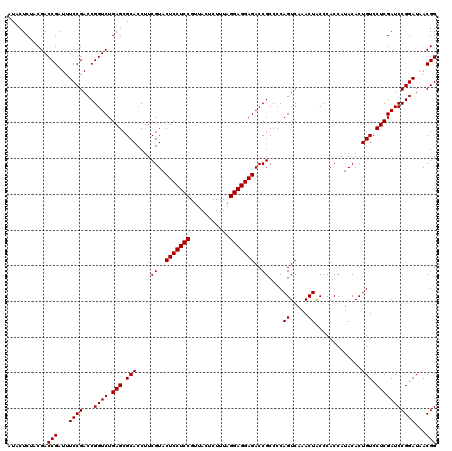
| Location | 5,264,940 – 5,265,060 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.13 |
| Consensus MFE | -40.13 |
| Energy contribution | -40.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264940 120 - 6264404 CCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAU (((((.(((.......))).....)))))(((((.....(((((...)))))((((((...........))))))((((....)))))))))(((((((.....)))))))......... ( -40.40) >pau.2917 5536872 120 - 6537648 CCGUGAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((..(((((.((((..((.((.(.....).)).)))))))))))))))..((((((...........))))))((((....))))((((.(((((((.....)))))))...)))).. ( -40.40) >ppk.656 4852234 120 + 6181863 CCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((((..(.....(((((.(.(((.((((.((....)).))))..))).).))))).....)..))))))....((((....))))((((.(((((((.....)))))))...)))).. ( -41.50) >pel.714 4508902 120 + 5888780 CCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((((..(.....(((((.(.(((.((((.((....)).))))..))).).))))).....)..))))))....((((....))))((((.(((((((.....)))))))...)))).. ( -41.50) >pf5.2519 4810809 120 - 7074893 CCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((((..(.....(((((.(.(((.((((.((....)).))))..))).).))))).....)..))))))....((((....))))((((.(((((((.....)))))))...)))).. ( -41.50) >pfo.2296 4417373 120 - 6438405 CCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((((..(.....(((((.(.(((.((((.((....)).))))..))).).))))).....)..))))))....((((....))))((((.(((((((.....)))))))...)))).. ( -41.50) >consensus CCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAU ((((((..(.....(((((.(.(((.((((.((....)).))))..))).).))))).....)..))))))....((((....))))((((.(((((((.....)))))))...)))).. (-40.13 = -40.47 + 0.33)

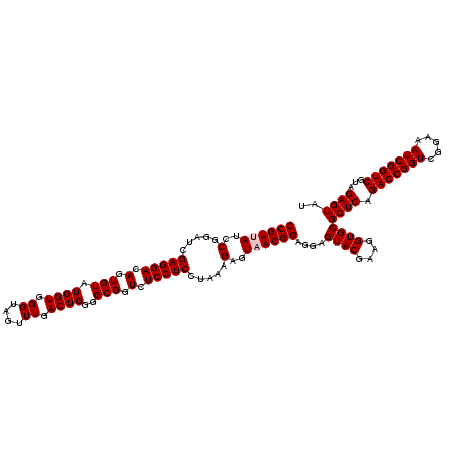
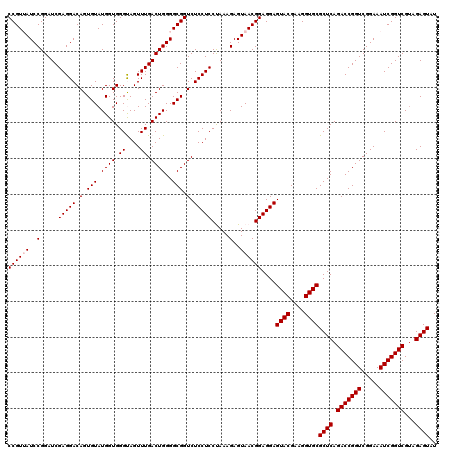
| Location | 5,264,980 – 5,265,100 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -33.73 |
| Energy contribution | -33.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264980 120 + 6264404 CGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUACGGACCAGAGUUAGAACCUCAAGCAUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))((((((((((.....)))...........(((..(((..((((.....))))...))).)))................)))))))...... ( -36.70) >pau.2917 5536912 120 + 6537648 CGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUCACGGACCAGAGUUAGAACCUCAAGCAUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))((((((((((.....)))...........(((..(((..((((.....))))...))).)))................)))))))...... ( -36.70) >ppk.656 4852274 120 - 6181863 CGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))...... ( -34.90) >pel.714 4508942 120 - 5888780 CGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAGGUUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))(((((((..((..................(((..((((.((((.....))))..)))).)))((((....))))))..)))))))...... ( -38.30) >pf5.2519 4810849 120 + 7074893 CGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))...... ( -35.50) >pfo.2296 4417413 120 + 6438405 CGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))...... ( -35.50) >consensus CGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCA ....(((((((...........)))))))(((((((((((.................))))..((((.((((.....)))).).)))....................)))))))...... (-33.73 = -33.73 + 0.00)

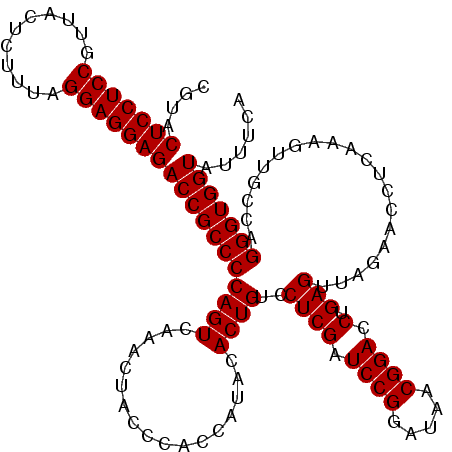
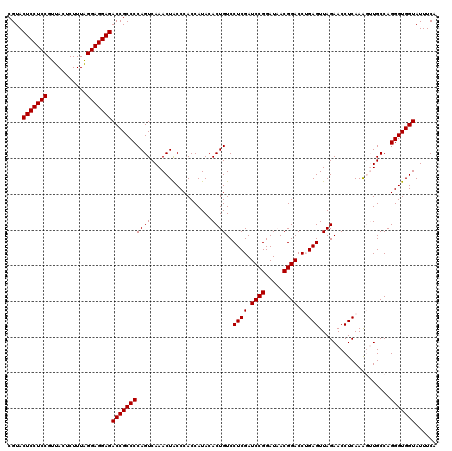
| Location | 5,265,020 – 5,265,140 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -42.05 |
| Consensus MFE | -41.85 |
| Energy contribution | -40.77 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.93 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265020 120 - 6264404 GGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUG (((((....((((((((....)))))))).)))))............(((((...((((.((((((.......)))(((((((.....))))))))))...))))..)))))........ ( -43.30) >pau.2917 5536952 120 - 6537648 GGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUGAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUG (((((....((((((((....)))))))).)))))............(((((...((((.((((((.......)))(((((((.....))))))))))...))))..)))))........ ( -43.30) >ppk.656 4852314 120 + 6181863 GGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUG (((((....((((((((....)))))))).)))))............(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))........ ( -40.40) >pel.714 4508982 120 + 5888780 GGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUG (((((....((((((((....)))))))).)))))............(((((...(((.(((((((.......))).((((((.....))))))))))....)))..)))))........ ( -43.00) >pf5.2519 4810889 120 - 7074893 GGCUUUGAAGCGUGAACGCCAGUUUGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUG (((((....(((..(((....)))..))).)))))............(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))........ ( -40.20) >pfo.2296 4417453 120 - 6438405 GGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUG (((((....((((((((....)))))))).)))))............(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))........ ( -42.10) >consensus GGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUG (((((....((((((((....)))))))).)))))............(((((...(((.(((((((.......))).((((((.....))))))))))....)))..)))))........ (-41.85 = -40.77 + -1.08)

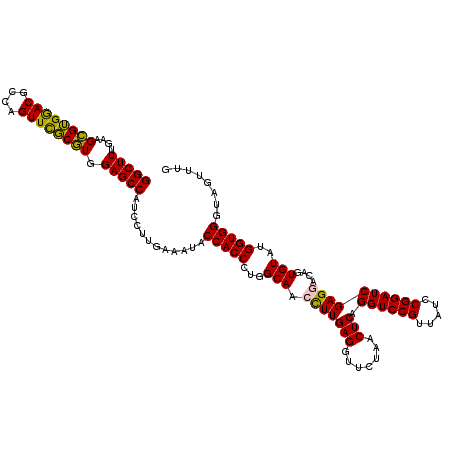
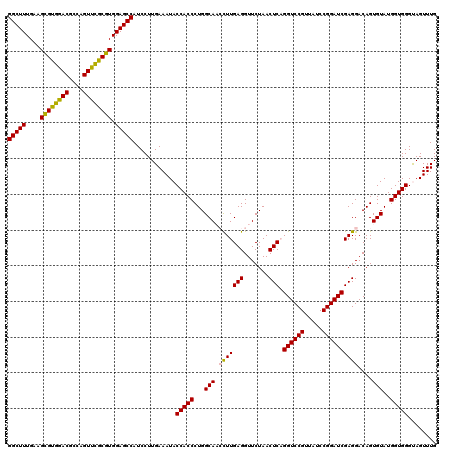
| Location | 5,265,060 – 5,265,180 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -42.48 |
| Energy contribution | -41.15 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5265060 120 - 6264404 UGCACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGU ......((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..)))))).......... ( -45.80) >pau.2917 5536992 120 - 6537648 UGCACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGU ......((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..)))))).......... ( -45.80) >ppk.656 4852354 120 + 6181863 UGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGU ......((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..)))))).......... ( -39.80) >pel.714 4509022 120 + 5888780 UGCACUGGACUUUGAGCUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGU ......((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..)))))).......... ( -41.60) >pf5.2519 4810929 120 - 7074893 UGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGAACGCCAGUUUGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGU ......((((((..((.(((((.(.((.((.((...(((((((((....(((..(((....)))..))).))))).)))).....)))))).).))))).))..)))))).......... ( -39.60) >pfo.2296 4417493 120 - 6438405 UGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGU ......((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..)))))).......... ( -41.50) >consensus UGCACUGGACUUUGAACUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGU ......((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..)))))).......... (-42.48 = -41.15 + -1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:15:14 2007