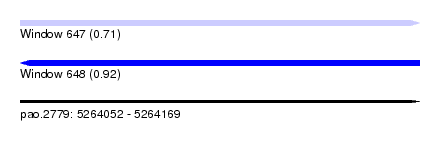
| Sequence ID | pao.2779 |
|---|---|
| Location | 5,264,052 – 5,264,169 |
| Length | 117 |
| Max. P | 0.918886 |

| Location | 5,264,052 – 5,264,169 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264052 117 + 6264404 CGCAAAAGC-AAAACCCCCGGU-CAUCGCU-GACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUU ((((...((-((((((((.(((-((....)-)))).))))))))((((((((..(........).)))).))))....(((....))).............))..))))........... ( -40.00) >pau.2917 5535985 117 + 6537648 CGCAAAAGC-AAAACCCCCGGC-CAUCGCU-GACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUU .((....))-........((((-(((((((-((....((....(((((((((..(........).)))).)))))...(((....))).......)).))))))))).))))........ ( -37.30) >pfo.3002 5726293 116 + 6438405 GGCAAAA-C-AAAACCCCAACUGCUUUCGC-AAUUGGGGUUUC-GGAAUUUGAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUU ((((((.-(-.(((((((((.(((....))-).))))))))).-)...)))).))..(((((((..............(((....)))........((((...)))))))))))...... ( -34.20) >pf5.3378 6382699 117 + 7074893 CGCAAAAAC-AAAACCCCAACUGCAUGCGC-AGUUGGGGUUUC-GGAAUUUAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUU ........(-.(((((((((((((....))-))))))))))).-)............(((((((..............(((....)))........((((...)))))))))))...... ( -39.10) >pst.2031 3869387 119 + 6397126 CAUAAAAACAAAAACCCUCGACGCGUAAGC-GAUGAGGGUUUUUGGAAUUUAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUACAUCGUUUCACUG ........((((((((((((.(((....))-).))))))))))))............(((((((..............(((....)))..(.((......)).)...)))))))...... ( -40.00) >pel.69 5768079 115 - 5888780 -----AGACAAAACCCCUACCUGCAUGCGCAGAUAGGGGUUUUGCGAAAUGAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUG -----.(.(((((((((((.((((....)))).))))))))))))(((((((...(((.(((((..............(((....)))........))))).)))....))))))).... ( -41.85) >consensus CGCAAAAAC_AAAACCCCAGCUGCAUCCGC_GAUCAGGGUUUUUGGAAUUGAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUU ..............((((.................))))..................(((((((..............(((....)))........((((...)))))))))))...... (-18.59 = -18.68 + 0.09)

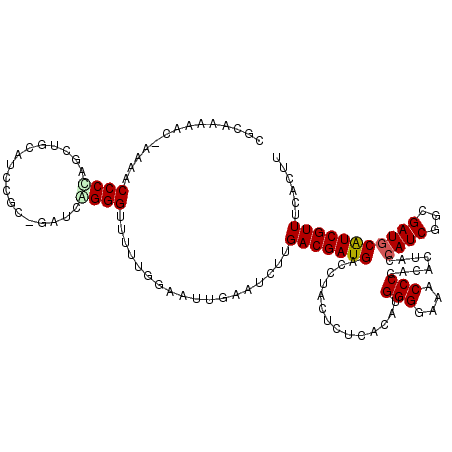
| Location | 5,264,052 – 5,264,169 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -47.97 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2779 5264052 117 - 6264404 AAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUC-AGCGAUG-ACCGGGGGUUUU-GCUUUUGCG ..(..(((.((((.((.(((.((....(..(((((...)))))..)....)).))).))))))............((((((((((((((-(....))-))))))))))))-).)))..). ( -49.70) >pau.2917 5535985 117 - 6537648 AAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUC-AGCGAUG-GCCGGGGGUUUU-GCUUUUGCG ..(..(((.((((.((.(((.((....(..(((((...)))))..)....)).))).))))))............((((((((((((((-(....))-))))))))))))-).)))..). ( -49.10) >pfo.3002 5726293 116 - 6438405 AAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCAAAUUCC-GAAACCCCAAUU-GCGAAAGCAGUUGGGGUUUU-G-UUUUGCC ..(..((((((((.((.(((.((....(..(((((...)))))..)....)).))).)))))).............-((((((((((((-((....))))))))))))))-)-)))..). ( -50.00) >pf5.3378 6382699 117 - 7074893 AAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUAAAUUCC-GAAACCCCAACU-GCGCAUGCAGUUGGGGUUUU-GUUUUUGCG ..(..((((((((....(((.((....(..(((((...)))))..)....)).))))))))).............(-((((((((((((-((....))))))))))))))-)..))..). ( -47.40) >pst.2031 3869387 119 - 6397126 CAGUGAAACGAUGUAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUAAAUUCCAAAAACCCUCAUC-GCUUACGCGUCGAGGGUUUUUGUUUUUAUG ..(((((((((((....(((.((....(..(((((...)))))..)....)).))))))))).............(((((((((((..(-((....)))..)))))))))))..))))). ( -41.50) >pel.69 5768079 115 + 5888780 CAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUCAUUUCGCAAAACCCCUAUCUGCGCAUGCAGGUAGGGGUUUUGUCU----- .((((((((((((....(((.((....(..(((((...)))))..)....)).))))))))).....))))))..(((((((((((((((((....)))))))))))))))))..----- ( -50.10) >consensus AAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCAAAUUCCAAAAACCCCAAUC_GCGCAUGCAGCGGGGGUUUU_GUUUUUGCG ..((((((.((((.((.(((.((....(..(((((...)))))..)....)).))).))))))..................((((.....((....))....)))).......)))))). (-23.28 = -24.23 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:58 2007