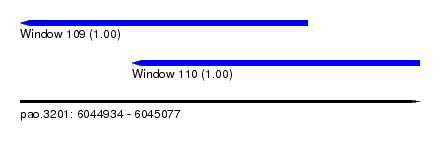
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,044,934 – 6,045,077 |
| Length | 143 |
| Max. P | 0.999889 |

| Location | 6,044,934 – 6,045,037 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.35 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
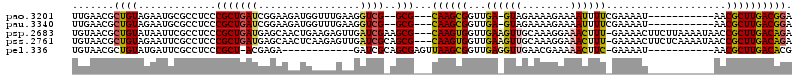
>pao.3201 6044934 103 - 6264404 GUUUGAAGGUCG--GCG---CAAGCGGUUGA-GUAGAAAAGAAAAUUUUCGAAAAU-----------AACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACG ((((.((..(((--.((---((((((.....-...(((((.....)))))......-----------..)))))).))...)))((((((((.((.....))))))))))..)).)))). ( -32.89) >pau.3340 6317853 103 - 6537648 GUUUGAAGGUCG--GCG---CAAGCGGUUGA-GUAGAAAAGAAAAUUUUCGAAAAU-----------AACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACG ((((.((..(((--.((---((((((.....-...(((((.....)))))......-----------..)))))).))...)))((((((((.((.....))))))))))..)).)))). ( -32.89) >psp.2683 5252413 115 - 5928787 AAGAGUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUU-GAAAACUUCUUAAAAUAACCGCUUGACAGAUACAAGAGACGCUGUAGAAUGCGCG-CCUCGGUUGAGACG .....(..(((((.(((---(((((((((((((((.(((((....))))-)..)))))).......))))))))(.((..((((........))))...))))))-).)))))..).... ( -39.51) >pss.2761 5428443 115 - 6093698 AAGAGUUGAUCGCAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUU-GAAAACUUCUCAAAAUAACCGCUUGACAGAUACAGAGGACGCUGUAGAAUGCGCG-CCUCGGUUGAGACG .....(..((((..(((---(((((((((((((((.(((((....))))-)..)))))).......))))))))(.((..(((((......)))))...))))))-)..))))..).... ( -40.21) >pel.336 5192459 100 + 5888780 -------GAUCGCAGCGAGUUAAGCGGUUGAGGUUGAACGAAAAACUUC-GAAAAU-----------AACGCUUGACACGAAAUGAGGUUAGCGUAGAAUGCGCG-CCUCGGUUGAGACG -------..((.((((..((((((((.((((((((........))))))-))....-----------..))))))))......((((((..((((.....)))))-))))))))).)).. ( -36.00) >consensus AAGAGAAGAUCG_AGCG___CAAGCGGUUGA_GUUGAAAAGAAAACUUU_GAAAAU___________AACGCUUGACAGAAACAGAGGUUGCUGUAGAAUGCGCG_CCUCGGUUGAGACG .........((...((....((((((...((((((........))))))....................)))))).........((((.(((.((.....))))).)))).))...)).. (-12.70 = -13.02 + 0.32)
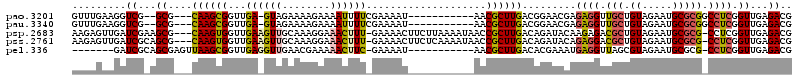
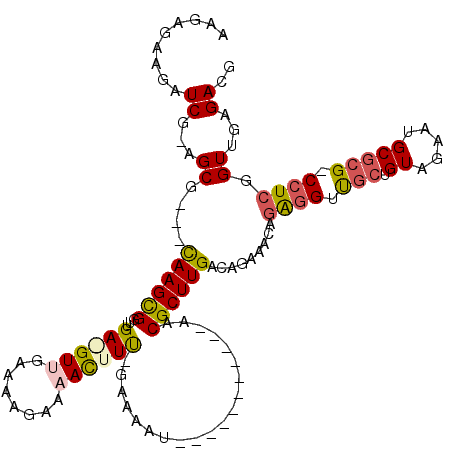
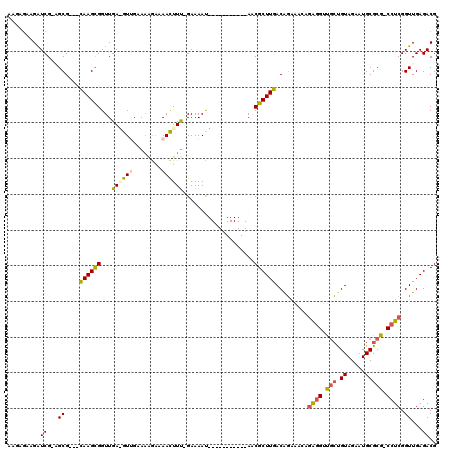
| Location | 6,044,974 – 6,045,077 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.48 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -9.27 |
| Energy contribution | -9.39 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.27 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6044974 103 - 6264404 UUGAACGCUGUAGAAUGCGCCUCCCGCUGAUCGGAAGAUGGUUUGAAGGUCG--GCG---CAAGCGGUUGA-GUAGAAAAGAAAAUUUUCGAAAAU-----------AACGCUUGACGGA .......((((.....(((((..(((((.(((....)))))).....))..)--)))---)(((((.....-...(((((.....)))))......-----------..))))).)))). ( -29.79) >pau.3340 6317893 103 - 6537648 UUGAACGCUGUAGAAUGCGCCUCCCGCUGAUCGGAAGAUGGUUUGAAGGUCG--GCG---CAAGCGGUUGA-GUAGAAAAGAAAAUUUUCGAAAAU-----------AACGCUUGACGGA .......((((.....(((((..(((((.(((....)))))).....))..)--)))---)(((((.....-...(((((.....)))))......-----------..))))).)))). ( -29.79) >psp.2683 5252452 116 - 5928787 UGUAACGCUGUAUAAUUCGCCUCCCGCUGAUGAGCAACUGAAGAGUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUU-GAAAACUUCUUAAAAUAACCGCUUGACAGA .......((((.............((((..(((.(((((....))))).))).))))---(((((((((((((((.(((((....))))-)..)))))).......))))))))))))). ( -38.31) >pss.2761 5428482 116 - 6093698 UGUAACGCUGUAGAAUUCGCCUCCCGCUGAUGAGCAACUCAAGAGUUGAUCGCAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUU-GAAAACUUCUCAAAAUAACCGCUUGACAGA .......((((.............(((((.(((.(((((....))))).))))))))---(((((((((((((((.(((((....))))-)..)))))).......))))))))))))). ( -42.11) >pel.336 5192498 95 + 5888780 UGUAACGCUGUAUGAUUCGCCUCCCGCU-ACGAGA------------GAUCGCAGCGAGUUAAGCGGUUGAGGUUGAACGAAAAACUUC-GAAAAU-----------AACGCUUGACACG .....((((((..(((.(..(((.....-..))).------------)))))))))).((((((((.((((((((........))))))-))....-----------..))))))))... ( -30.50) >consensus UGUAACGCUGUAGAAUUCGCCUCCCGCUGAUGAGAAAAUGAAGAGAAGAUCG_AGCG___CAAGCGGUUGA_GUUGAAAAGAAAACUUU_GAAAAU___________AACGCUUGACAGA .......((((.............(((((((.................))))..)))...((((((...((((((........))))))....................)))))))))). ( -9.27 = -9.39 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:15 2007