
| Sequence ID | pao.1087 |
|---|---|
| Location | 1,954,710 – 1,954,782 |
| Length | 72 |
| Max. P | 0.991229 |

| Location | 1,954,710 – 1,954,782 |
|---|---|
| Length | 72 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.38 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1087 1954710 72 + 6264404 GGCUCCGCAAUACUGGCGGUCGAGACAUUCGGCCGCCUCGUGGGCUUGAAAAUGCCCACAAUUGCCUUCAUC ......(((((...((((((((((...))))))))))..((((((........)))))).)))))....... ( -34.00) >psp.1009 1962855 63 + 5928787 --------AAUGUAGGGGGUCGGCACAACCGACCGCCU-GCGGACUUGAAAAAGCCCGCAAUAGCCUUCAUC --------.....(((.((((((.....)))))).)))-((((.(((....))).))))............. ( -27.30) >pst.2209 2188904 63 - 6397126 --------AAUGUAGGGGGUCGGCACAACCGACCGCCU-GCGGACUUGAAAAAGCCCGCAAUAGCCUUCAUC --------.....(((.((((((.....)))))).)))-((((.(((....))).))))............. ( -27.30) >pf5.2419 2469150 63 - 7074893 --------AAUAGGCAUGGUCGGUUGUUCCGAUCAGUG-GCGGGCUUGAAAAAGCCCGCAAUAGCCUUCAUC --------...((((.(((((((.....)))))))...-((((((((....))))))))....))))..... ( -33.30) >pfo.2166 2252652 63 - 6438405 --------AAUAGGCUUGGUCGGCGCGUCUGAUCAGCG-GCGGGCUUGAAAAAGCCCGCAAUAGCCUUCAUC --------...((((((((((((.....)))))))...-((((((((....))))))))...)))))..... ( -30.00) >pss.1035 1985433 63 + 6093698 --------AAUGUAGGGGGUCGGCACAACCGACCGCCU-GCGGACUUGAAAAAGCCCGCAAUAGCCUUCAUC --------.....(((.((((((.....)))))).)))-((((.(((....))).))))............. ( -27.30) >consensus ________AAUAUAGGGGGUCGGCACAACCGACCGCCU_GCGGACUUGAAAAAGCCCGCAAUAGCCUUCAUC ..............((.((((((.....)))))).))..((((.(((....))).))))............. (-19.82 = -20.72 + 0.89)

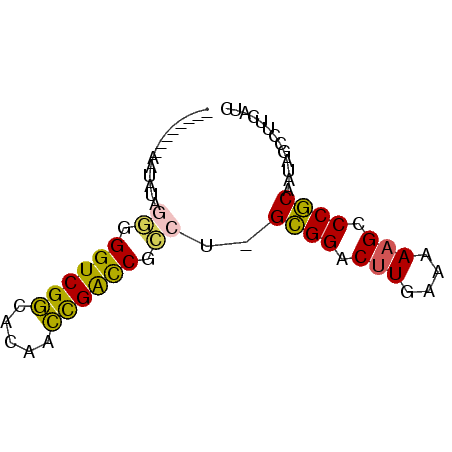
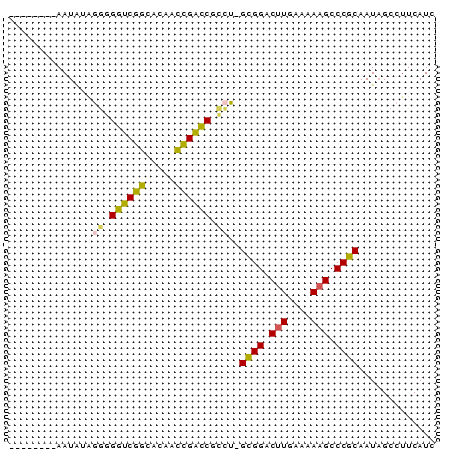
| Location | 1,954,710 – 1,954,782 |
|---|---|
| Length | 72 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.13 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1087 1954710 72 - 6264404 GAUGAAGGCAAUUGUGGGCAUUUUCAAGCCCACGAGGCGGCCGAAUGUCUCGACCGCCAGUAUUGCGGAGCC ......(((..((((((((........))))))))(((((.(((.....))).)))))...........))) ( -31.30) >psp.1009 1962855 63 - 5928787 GAUGAAGGCUAUUGCGGGCUUUUUCAAGUCCGC-AGGCGGUCGGUUGUGCCGACCCCCUACAUU-------- ((((.(((...((((((((((....))))))))-))..((((((.....)))))).))).))))-------- ( -29.60) >pst.2209 2188904 63 + 6397126 GAUGAAGGCUAUUGCGGGCUUUUUCAAGUCCGC-AGGCGGUCGGUUGUGCCGACCCCCUACAUU-------- ((((.(((...((((((((((....))))))))-))..((((((.....)))))).))).))))-------- ( -29.60) >pf5.2419 2469150 63 + 7074893 GAUGAAGGCUAUUGCGGGCUUUUUCAAGCCCGC-CACUGAUCGGAACAACCGACCAUGCCUAUU-------- .....((((....((((((((....))))))))-...((.((((.....)))).)).))))...-------- ( -24.80) >pfo.2166 2252652 63 + 6438405 GAUGAAGGCUAUUGCGGGCUUUUUCAAGCCCGC-CGCUGAUCAGACGCGCCGACCAAGCCUAUU-------- .....(((((...((((((((....))))))))-(((.........))).......)))))...-------- ( -22.90) >pss.1035 1985433 63 - 6093698 GAUGAAGGCUAUUGCGGGCUUUUUCAAGUCCGC-AGGCGGUCGGUUGUGCCGACCCCCUACAUU-------- ((((.(((...((((((((((....))))))))-))..((((((.....)))))).))).))))-------- ( -29.60) >consensus GAUGAAGGCUAUUGCGGGCUUUUUCAAGCCCGC_AGGCGGUCGGAUGUGCCGACCCCCUACAUU________ ((((...(((...((((((((....))))))))..)))((((((.....)))))).....))))........ (-20.18 = -20.13 + -0.05)

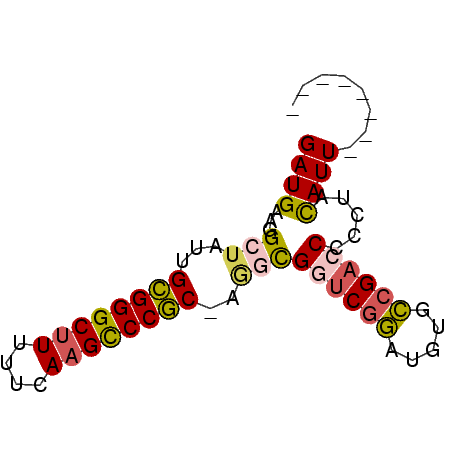
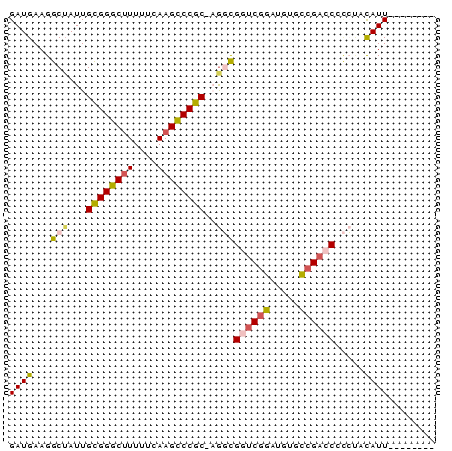
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:41 2007