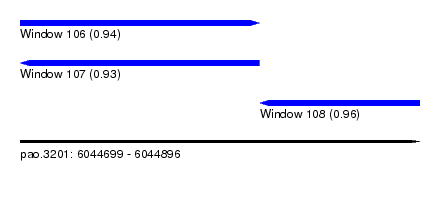
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,044,699 – 6,044,896 |
| Length | 197 |
| Max. P | 0.964154 |

| Location | 6,044,699 – 6,044,817 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
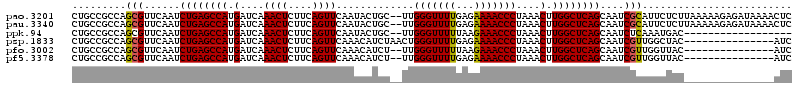
>pao.3201 6044699 118 + 6264404 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUC .........(((......((((((((.(...........((((.....)))).--..((((((.....))))))....).))))))))....)))..(((((.....)))))........ ( -30.90) >pau.3340 6317618 118 + 6537648 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAUUCUCUUAAAAAGAGAUAAAACUC .........(((......((((((((.(...........((((.....)))).--..((((((.....))))))....).))))))))....)))..(((((.....)))))........ ( -30.90) >ppk.94 6005017 100 - 6181863 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAUACUGC--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCUCAAAUGAC------------------ ..((.....)).......((((((((.(...........((((.....)))).--..(((((((...)))))))....).))))))))..............------------------ ( -23.40) >psp.1833 3636649 105 + 5928787 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCUAACUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGUUGGCUAC---------------AUC .....(((((((......((((((((.(....((((....)))).............((((((.....))))))....).))))))))....)))))))...---------------... ( -32.50) >pfo.3002 5731515 103 + 6438405 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCU--UUGGGUUUUUAAGAAACCCUAAACUUGGCUCAGCAAUCGUUGGUUAC---------------AUC .((..(((((((......((((((((.(....((((....)))).........--..(((((((...)))))))....).))))))))....)))))))..)---------------).. ( -30.90) >pf5.3378 6387908 103 + 7074893 CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAACAUCU--UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGUUGGUUAC---------------AUC .((..(((((((......((((((((.(....((((....)))).........--..((((((.....))))))....).))))))))....)))))))..)---------------).. ( -30.50) >consensus CUGCCGCCAGCGUUCAAUCUGAGCCAUGAUCAAACUCUUCAGUUCAAAAAUCC__UUGGGUUUUGAGAAAACCCUAAACUUGGCUCAGCAAUCGCAGGCUAC_______________AUC .........(((......((((((((.(....((((....)))).............(((((((...)))))))....).))))))))....)))......................... (-22.42 = -22.12 + -0.31)
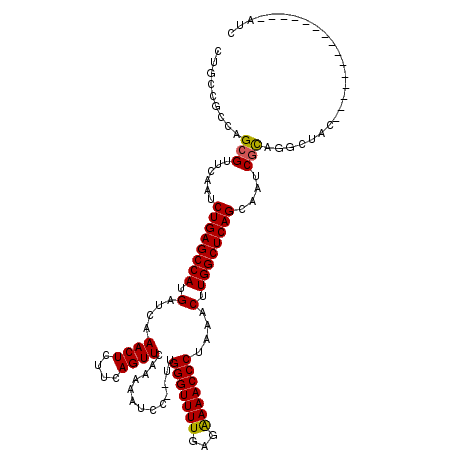
| Location | 6,044,699 – 6,044,817 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6044699 118 - 6264404 GAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ........(((((.....))))).(((.....((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))......((....))))). ( -37.60) >pau.3340 6317618 118 - 6537648 GAGUUUUAUCUCUUUUUAAGAGAAUGCGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ........(((((.....))))).(((.....((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).))))))))......((....))))). ( -37.60) >ppk.94 6005017 100 + 6181863 ------------------GUCAUUUGAGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--GCAGUAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ------------------((((((..(.....((((((((.((((.((((((.....))))))))--)).(.((..((((....))))..))).)))))))).)..))...))))..... ( -32.60) >psp.1833 3636649 105 - 5928787 GAU---------------GUAGCCAACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAGUUAGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ..(---------------((.((((.((.((.(((((((.......((((((.....))))))......(((.(..((((....))))..).))))))))))...)).)).)))).))). ( -37.80) >pfo.3002 5731515 103 - 6438405 GAU---------------GUAACCAACGAUUGCUGAGCCAAGUUUAGGGUUUCUUAAAAACCCAA--AGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ..(---------------((..(((.((.((.(((((((.......((((((.....))))))..--..(((.(..((((....))))..).))))))))))...)).)).)))..))). ( -34.60) >pf5.3378 6387908 103 - 7074893 GAU---------------GUAACCAACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA--AGAUGUUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ..(---------------((..(((.((.((.(((((((.......((((((.....))))))..--..(((.(..((((....))))..).))))))))))...)).)).)))..))). ( -34.70) >consensus GAU_______________GUAACCAACGAUUGCUGAGCCAAGUUUAGGGUUUUCUCAAAACCCAA__ACAGGAUUGAACUGAAGAGUUUGAUCAUGGCUCAGAUUGAACGCUGGCGGCAG ................................((((((((......((((((.....))))))..........(..((((....))))..)...)))))))).......((.....)).. (-27.12 = -27.12 + -0.00)

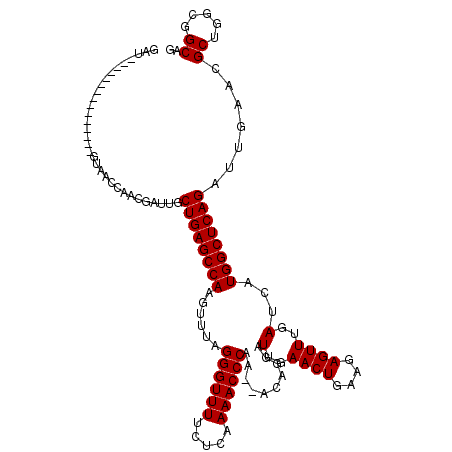
| Location | 6,044,817 – 6,044,896 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6044817 79 - 6264404 GCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCGCAA-GAUUAUCAGCAACGCAAGUAACACU-CGUGAAUUCGA (.((((((.(((..((((((((.((.....(((((((((....-))))))))))).)))))))).))).-..))))))).. ( -29.40) >pau.3340 6317736 79 - 6537648 GCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUCGCAA-GAUUAUCAGCAACGCAAGUAACACU-CGUGAAUUCGA (.((((((.(((..((((((((.((.....(((((((((....-))))))))))).)))))))).))).-..))))))).. ( -29.40) >ppk.94 6005120 79 + 6181863 --AAUUCGUGUGGGUGCUUGUGAGUACGGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUGGCCAUGCGAGAAAUCAC --..(((((((((.(((((((((.....(.(((((((((.....)))))))))).))))))))).)))))))))....... ( -36.40) >psp.1833 3636755 77 - 5928787 --AAUUCGUGUGGGUGCUUGUGGUGUCAGACUGA-AGUCAACA-GAUUAUCAGCAACCCAAGUUACUCCGCGAGAAAUCAA --..((((((.(((((((((.(((....(.((((-((((....-)))).))))).))))))).)))))))))))....... ( -25.70) >pf5.3378 6388012 81 - 7074893 GCAAUUCGUGUGGGUGCUUGUGGAGUCAGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUUACUCCGCGAGAAAUCAA ....((((((.(((((((((((..(.....(((((((((.....))))))))))..)))))).)))))))))))....... ( -30.10) >pst.2031 3874743 79 - 6397126 GCAAUUCGUGUGGGUGCUUGUGGUGUCAGACUGA-AGUCAACA-GAUUAUCAGCAACCCAAGUUACUCCGCGAGAAAUCAA ....((((((.(((((((((.(((....(.((((-((((....-)))).))))).))))))).)))))))))))....... ( -25.70) >consensus GCAAUUCGUGUGGGUGCUUGUGAUGUCAGACUGAUAGUCAAAA_GAUUAUCAGCAACACAAGUUACUCCGCGAGAAAUCAA ....(((.((((((((((((((......(.(((((((((.....))))))))))..))))))).)).))))).)))..... (-21.10 = -21.10 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:05:13 2007