| Sequence ID | pao.1203 |
|---|---|
| Location | 2,185,395 – 2,185,502 |
| Length | 107 |
| Max. P | 0.999152 |

| Location | 2,185,395 – 2,185,502 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.96 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
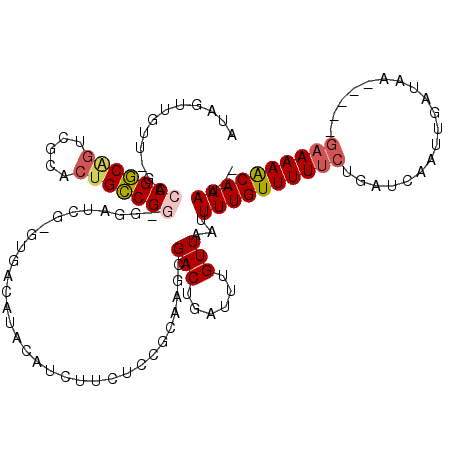
>pao.1203 2185395 107 + 6264404 UUAUUUGUUUUUC-----UCAUCAAUGGAUUGGAAAAACAAACUUAACAAAUCAGUCCGCAGCGGAGAAGCUCUGUGCGAC--------CCGGCAGUCAGACUGCCGCACGCACAAUCAC ...((((((((((-----(.(((....))).))))))))))).............((((...)))).......((((((..--------.((((((.....))))))..))))))..... ( -34.90) >pau.1822 3088856 107 - 6537648 UUAUUUGUUUUUC-----UCAUCAAUGGAUUGGAAAAACAAACUUAACAAAUCAGUCCGCAGCGGAGAAGCUCUGUGCGAC--------CCGGCAGUCAGACUGCCGCACGCACAAUCAC ...((((((((((-----(.(((....))).))))))))))).............((((...)))).......((((((..--------.((((((.....))))))..))))))..... ( -34.90) >psp.1845 3659441 112 + 5928787 --AUUUGUUUUUC-----UUAUCAAUUGAUCAGAAAAACAAAAUUAACAAAUCAGUCGCUGCGAGAGAAGAUGCAAGUCACCCGAUCCUCCGACAGUGCGACUGUCGGUC-AACAACUAU --.((((((((((-----(.(((....))).)))))))))))........(((.((.((((((........))).))).))..)))...(((((((.....)))))))..-......... ( -28.40) >pst.2042 3897482 112 + 6397126 --AUUUGCUUUUC-----UUAUCAAUCAAUCAGAAAAACAAAAUUAACAAAUCAGUCUCAUCAGGAGAAGAUGCACAUCAAGCCAUCCUCCGGCGGCGCGACUGCCGGUC-AACAACUAU --.((((.(((((-----(.((......)).)))))).)))).............((((.....)))).(((((.......).))))..(((((((.....)))))))..-......... ( -21.90) >pss.1932 3875499 112 + 6093698 --AUUUGCUUUUC-----UUAUGAAUUGAUCAGAAAAACAAAAUUAACAAAUCAGUCGCACCGAGAGAAGAUGCAGGUCACCCGAUCCUCCGGCAGCGCGACUGCCGGUC-AACAACUAU --.....((((((-----((.((.((((((....................))))))..))..)))))))).....((((....))))..(((((((.....)))))))..-......... ( -24.05) >ppk.1794 3533192 112 + 6181863 ---UUUGUUUUGUAAAAAUUAUCAAUCGAUGACGAAAACAAAAUUAACAAAUCAGUCGCUUGCGGUGAAGAUGUCUG-CACACGCUCA-CCGACAGC---CUCGUCGGUUAAACAACUAC ---...(((((((.....(((((....))))).....))))))).........(((.....((((((.((....)).-))).)))..(-(((((...---...))))))......))).. ( -22.90) >consensus __AUUUGUUUUUC_____UUAUCAAUCGAUCAGAAAAACAAAAUUAACAAAUCAGUCGCAACCGGAGAAGAUGCAUGUCAC_CGAUCC_CCGGCAGCGCGACUGCCGGUC_AACAACUAC ...((((((((((.....((.......))...))))))))))...............................................(((((((.....)))))))............ (-13.73 = -13.85 + 0.12)


| Location | 2,185,395 – 2,185,502 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.96 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.42 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1203 2185395 107 - 6264404 GUGAUUGUGCGUGCGGCAGUCUGACUGCCGG--------GUCGCACAGAGCUUCUCCGCUGCGGACUGAUUUGUUAAGUUUGUUUUUCCAAUCCAUUGAUGA-----GAAAAACAAAUAA ....((((((((.((((((.....)))))).--------).)))))))(((.((((((...))))..))...)))..(((((((((((..(((....)))..-----))))))))))).. ( -37.70) >pau.1822 3088856 107 + 6537648 GUGAUUGUGCGUGCGGCAGUCUGACUGCCGG--------GUCGCACAGAGCUUCUCCGCUGCGGACUGAUUUGUUAAGUUUGUUUUUCCAAUCCAUUGAUGA-----GAAAAACAAAUAA ....((((((((.((((((.....)))))).--------).)))))))(((.((((((...))))..))...)))..(((((((((((..(((....)))..-----))))))))))).. ( -37.70) >psp.1845 3659441 112 - 5928787 AUAGUUGUU-GACCGACAGUCGCACUGUCGGAGGAUCGGGUGACUUGCAUCUUCUCUCGCAGCGACUGAUUUGUUAAUUUUGUUUUUCUGAUCAAUUGAUAA-----GAAAAACAAAU-- .((((((((-(.(((((((.....))))))).(((..(((((.....)))))..)))..)))))))))..........(((((((((((.(((....))).)-----)))))))))).-- ( -40.10) >pst.2042 3897482 112 - 6397126 AUAGUUGUU-GACCGGCAGUCGCGCCGCCGGAGGAUGGCUUGAUGUGCAUCUUCUCCUGAUGAGACUGAUUUGUUAAUUUUGUUUUUCUGAUUGAUUGAUAA-----GAAAAGCAAAU-- ......(((-(((..((((((.((.((..((((((((((.....)).))))))))..)).)).))))).)..))))))(((((((((((.(((....))).)-----)))))))))).-- ( -32.80) >pss.1932 3875499 112 - 6093698 AUAGUUGUU-GACCGGCAGUCGCGCUGCCGGAGGAUCGGGUGACCUGCAUCUUCUCUCGGUGCGACUGAUUUGUUAAUUUUGUUUUUCUGAUCAAUUCAUAA-----GAAAAGCAAAU-- ......(((-(((..((((((((((((..((((((((((....)).).)))))))..))))))))))).)..))))))(((((((((((............)-----)))))))))).-- ( -45.30) >ppk.1794 3533192 112 - 6181863 GUAGUUGUUUAACCGACGAG---GCUGUCGG-UGAGCGUGUG-CAGACAUCUUCACCGCAAGCGACUGAUUUGUUAAUUUUGUUUUCGUCAUCGAUUGAUAAUUUUUACAAAACAAA--- .(((((((((.(((((((..---..))))))-)..(((.(((-.((....)).))))))))))))))).(((((.((..(((((.(((....)))..)))))..)).))))).....--- ( -34.50) >consensus AUAGUUGUU_GACCGGCAGUCGCACUGCCGG_GGAUCG_GUGACAUACAUCUUCUCCGCAAGCGACUGAUUUGUUAAUUUUGUUUUUCUGAUCAAUUGAUAA_____GAAAAACAAAU__ ............(((((((.....)))))))................................(((......)))...((((((((((...................))))))))))... (-16.15 = -16.38 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:35 2007