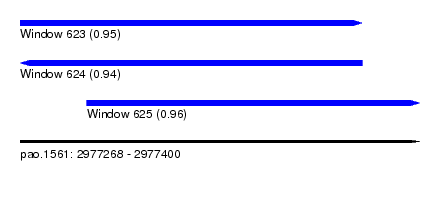
| Sequence ID | pao.1561 |
|---|---|
| Location | 2,977,268 – 2,977,400 |
| Length | 132 |
| Max. P | 0.963481 |

| Location | 2,977,268 – 2,977,381 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.48 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1561 2977268 113 + 6264404 GACCAUCC----GGUCACUCUUA-GAC-CUUCGUGUAGUAGUCUUGAAAAAACACUACAAAACGUUUGAC-GACGGAUUUUUUCCAGAGCAUGAUGGCCCCGCUCCCGCUAAUCAGGGGC ((((....----)))).((((((-(((-.....((((((.((.((....))))))))))....)))))).-...(((.....))).))).......(((((..............))))) ( -29.24) >pss.1912 3825429 113 + 6093698 GACUGGCC----GGUCG-ACUUCUCAC-AAAUAUGUAGUGAUUUAAAAAAAGCACUACAAAACGUUUGAC-GCCCCCAGAAGUGCGAGGCAUGAUUGCCUCGCUCCCGCUAAUCAGAGGC ((.((((.----((...-((((((...-.....(((((((.(((....))).)))))))...((.....)-).....))))))((((((((....)))))))).)).)))).))...... ( -37.10) >pfo.2115 4070631 113 + 6438405 GACUGAUU----AGUCG-UAUUA-GACGAAAGAUGUAGUGCUUGAAAAAAAGCACUACAAAACGUUUGAC-GAGCCAAAAAAGCGAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGC ..((((((----((.((-..(((-((((.....((((((((((......))))))))))...))))))).-..........(((((.(((......))))))))..)))))))))).... ( -42.60) >pf5.2365 4496836 113 + 7074893 GACUGAUU----AGUCG-UAUUA-GACCAAAUAUGUAGUGCCGCAAAAAAAGCACUACAAAACAUUUGAC-GAGCCAAAAAACCGCGGGCAUGAUGGCCUCGCUCCCGCUAAUCAGGGGC ..((((((----((.((-.....-...(((((.((((((((..........))))))))....)))))..-.............((((((......))).)))...)))))))))).... ( -33.20) >psp.1823 3605972 112 + 5928787 GACUCACC----AGUCU-AACUA-CGC-AAUUAUGUAGUGAGAUUAAAGAAGCACUACAAAUUGUUUGAC-GACCCCGAUACCCCGAGGCAUGAUUGCCUCGCUCCCGCUGAUCGGAGGC ((((....----)))).-.....-.((-((((.(((((((...((....)).))))))))))))).....-..(((((((.....((((((....))))))((....))..))))).)). ( -34.00) >pel.2001 3757331 117 + 5888780 -ACUGACCGCAGAGUCG-CAUUA-GACCAAAGAUGUAGUAAGCGCAAAAAAGCACUACAAAACAUUUGACAGGCUCUUUUUGGCAAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGC -.((((..((((((((.-..(((-((.......((((((..((........)))))))).....)))))..))))))....(((..((((......)))).)))...))...)))).... ( -27.30) >consensus GACUGACC____AGUCG_UAUUA_GAC_AAAUAUGUAGUGAUCUAAAAAAAGCACUACAAAACGUUUGAC_GACCCAAAAAACCCAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGC ((((........)))).................(((((((............)))))))...(.(((((.................((((......)))).((....))...))))).). (-16.92 = -16.37 + -0.55)

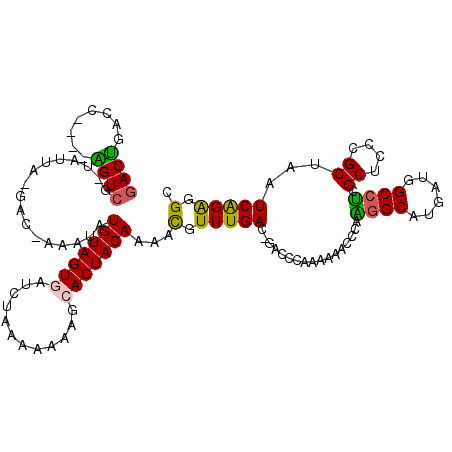
| Location | 2,977,268 – 2,977,381 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.48 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -19.63 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1561 2977268 113 - 6264404 GCCCCUGAUUAGCGGGAGCGGGGCCAUCAUGCUCUGGAAAAAAUCCGUC-GUCAAACGUUUUGUAGUGUUUUUUCAAGACUACUACACGAAG-GUC-UAAGAGUGACC----GGAUGGUC ((.((((.....)))).))..(((((((.(((((((((.....)))...-.....((.(((((((((((((.....)))).)))))).))).-)).-..))))))...----.))))))) ( -33.20) >pss.1912 3825429 113 - 6093698 GCCUCUGAUUAGCGGGAGCGAGGCAAUCAUGCCUCGCACUUCUGGGGGC-GUCAAACGUUUUGUAGUGCUUUUUUUAAAUCACUACAUAUUU-GUGAGAAGU-CGACC----GGCCAGUC ......((((.((.((.((((((((....))))))))((((((.(((((-(.....))))))((((((.(((....))).))))))......-...))))))-...))----.)).)))) ( -40.50) >pfo.2115 4070631 113 - 6438405 GCCUCUGAUUAGCGGGAGCGAGGCCAUCAUGCCUUCGCUUUUUUGGCUC-GUCAAACGUUUUGUAGUGCUUUUUUUCAAGCACUACAUCUUUCGUC-UAAUA-CGACU----AAUCAGUC ....((((((((((((((((((((......))).))))))).((((...-.))))(((...((((((((((......)))))))))).....))).-.....-)).))----)))))).. ( -39.90) >pf5.2365 4496836 113 - 7074893 GCCCCUGAUUAGCGGGAGCGAGGCCAUCAUGCCCGCGGUUUUUUGGCUC-GUCAAAUGUUUUGUAGUGCUUUUUUUGCGGCACUACAUAUUUGGUC-UAAUA-CGACU----AAUCAGUC ....((((((((((...(((((.(((....(((...)))....))))))-))((((((...(((((((((........)))))))))))))))...-.....-)).))----)))))).. ( -39.40) >psp.1823 3605972 112 - 5928787 GCCUCCGAUCAGCGGGAGCGAGGCAAUCAUGCCUCGGGGUAUCGGGGUC-GUCAAACAAUUUGUAGUGCUUCUUUAAUCUCACUACAUAAUU-GCG-UAGUU-AGACU----GGUGAGUC (((.((((((.((....))((((((....))))))...).)))))((((-......((((((((((((............))))))).))))-)..-.....-.))))----)))..... ( -34.74) >pel.2001 3757331 117 - 5888780 GCCUCUGAUUAGCGGGAGCGAGGCCAUCAUGCCUUUGCCAAAAAGAGCCUGUCAAAUGUUUUGUAGUGCUUUUUUGCGCUUACUACAUCUUUGGUC-UAAUG-CGACUCUGCGGUCAGU- ....((((((.(((((.((.((((((.((.((((((.....)))).)).))....((((...(((((((......)))).))).))))...)))))-)...)-)...))))))))))).- ( -34.50) >consensus GCCUCUGAUUAGCGGGAGCGAGGCCAUCAUGCCUUGGACUAUUUGGGUC_GUCAAACGUUUUGUAGUGCUUUUUUUAAAUCACUACAUAUUU_GUC_UAAUA_CGACU____GGUCAGUC ((.((((.....)))).))(((((......)))))..........................(((((((............))))))).................((((........)))) (-19.63 = -18.97 + -0.66)

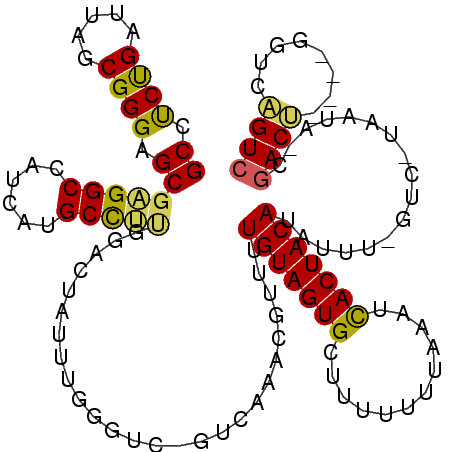
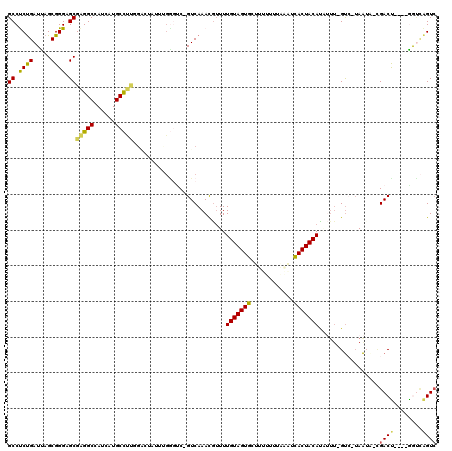
| Location | 2,977,290 – 2,977,400 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.51 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
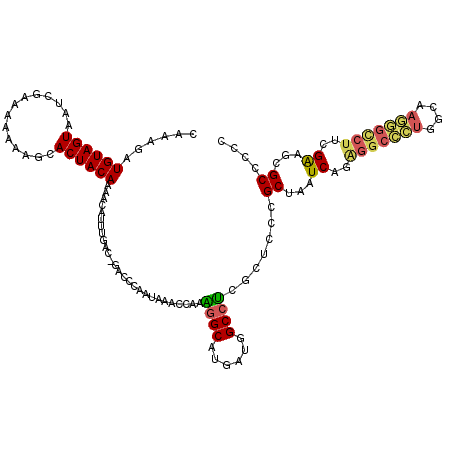
>pao.1561 2977290 110 + 6264404 -CUUCGUGUAGUAGUCUUGAAAAAACACUACAAAACGUUUGAC-GACGGAUUUUUUCCAGAGCAUGAUGGCCCCGCUCCCGCUAAUCAGGGGCUCUGAAA--------CGACGAGCUCCC -.((((((((((.((.((....)))))))))....(((((...-...(((.....)))(((((.(((((((.........))).))))...))))).)))--------)))))))..... ( -28.30) >pss.1912 3825451 118 + 6093698 -AAAUAUGUAGUGAUUUAAAAAAAGCACUACAAAACGUUUGAC-GCCCCCAGAAGUGCGAGGCAUGAUUGCCUCGCUCCCGCUAAUCAGAGGCUUUGCAAAAAGCCUUCGAGUCGCCGCU -.....(((((((.(((....))).)))))))........(((-......((..(.((((((((....)))))))).)...))..((..(((((((....)))))))..)))))...... ( -40.00) >pfo.2115 4070652 119 + 6438405 GAAAGAUGUAGUGCUUGAAAAAAAGCACUACAAAACGUUUGAC-GAGCCAAAAAAGCGAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGCCCUGGCAAGGGCCUUCGAAGCGCUGCU ......((((((((((......))))))))))...((.....)-)(((......(((((.(((......))))))))..((((..((..(((((((....)))))))..))))))..))) ( -49.00) >pf5.2365 4496857 119 + 7074893 CAAAUAUGUAGUGCCGCAAAAAAAGCACUACAAAACAUUUGAC-GAGCCAAAAAACCGCGGGCAUGAUGGCCUCGCUCCCGCUAAUCAGGGGCCCUGGCAAGGGCCUUCGGAUCGCCGCU (((((.((((((((..........))))))))....))))).(-((.((........(((((..(((.....)))..)))))......((((((((....)))))))).)).)))..... ( -41.00) >ppk.2374 4653090 120 + 6181863 CAAAGAUGUAGUAAGGACAAAAAAGCACUACAAAACAUUUGACAGGGUCUUUUUCCCAAGGGCAUGAUGGCCUCGCUCCCGCUAAUCAGGGGCCCUGGCAAGGGCCUUCGGGGCGCGCUC ((((..((((((..............)))))).....))))...(((.......)))..((((......))))((((((((.......((((((((....))))))))))))).)))... ( -36.95) >pel.2001 3757355 120 + 5888780 CAAAGAUGUAGUAAGCGCAAAAAAGCACUACAAAACAUUUGACAGGCUCUUUUUGGCAAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGCCCUGGCAAGGGUCUUCGGAGCGCCCUC ((((..((((((..((........)))))))).....)))).............(((..((((......)))).(((((.........((((((((....)))))))).))))))))... ( -36.10) >consensus CAAAGAUGUAGUAAUCGAAAAAAAGCACUACAAAACAUUUGAC_GACCCAAUAAACCAAAGGCAUGAUGGCCUCGCUCCCGCUAAUCAGAGGCCCUGGCAAGGGCCUUCGAAGCGCCCCC ......((((((..............))))))...........................((((......)))).......((...((..(((((((....)))))))..))...)).... (-21.53 = -21.51 + -0.02)

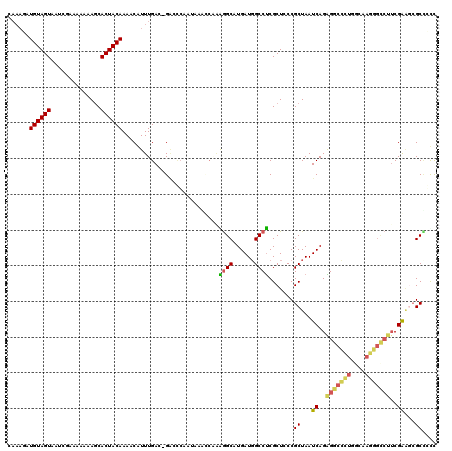
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:33 2007