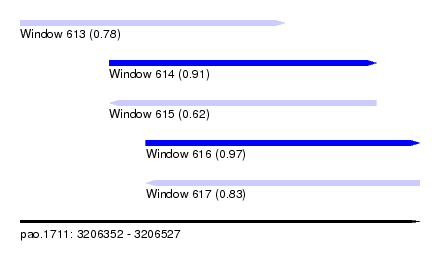
| Sequence ID | pao.1711 |
|---|---|
| Location | 3,206,352 – 3,206,527 |
| Length | 175 |
| Max. P | 0.967698 |

| Location | 3,206,352 – 3,206,468 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -25.93 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
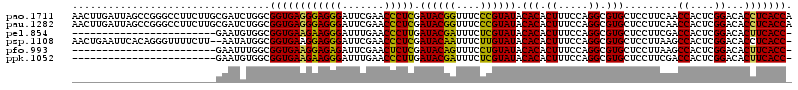
>pao.1711 3206352 116 + 6264404 CGGGAAAUGUUCGAGUAAAUU-GCGCGCCUCGACAGACACGGCGC---UGUCGAGAACCUGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUA ((((.....)))).......(-((((((((((((((........)---)))))))..((.((((..((.(((...........))))).)))).))...)))))))(((((....))))) ( -41.00) >pau.1282 4174618 116 - 6537648 CGGGAAAUGUUCGAGUAAAUU-GCGCGCCUCGACAGACACGGCGC---UGUCGAGAACCUGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUA ((((.....)))).......(-((((((((((((((........)---)))))))..((.((((..((.(((...........))))).)))).))...)))))))(((((....))))) ( -41.00) >pel.854 1616177 118 + 5888780 GAAAAGCUCUUCUACUAAAUUAGCGCGCCUCGACAGGCUGAAUAG-CUUGAAGAGAAAC-GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUA .....(((((((((((............(((..((((((....))-))))..)))...(-((........))))))))....))))))((((((....)).)))).(((((....))))) ( -37.10) >pf5.1100 2042029 119 + 7074893 UGAAAAGCGAUCGACUACAUUAGCGCGCCUCGACAGACUGAAUCGGUUUGAAGAGAUAC-GGUGAAGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAGGAAACUGUA ....((((.((((..(((.(((.((...(((..(((((((...)))))))..)))...)-).))).)))..))).).)))).....((((((.(....))))))).(((((....))))) ( -37.10) >pfo.993 1938114 119 + 6438405 AGAAAAGCUUUCGACUACAUUAGCGCGCCUCGACAGACAGAACAUGUUUGAAGAGAUAC-GGUGAAGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAGGAAACUGUA ....(((((((((..(((.(((.((...(((..((((((.....))))))..)))...)-).))).)))..)))).))))).....((((((.(....))))))).(((((....))))) ( -38.00) >ppk.1052 2069002 118 + 6181863 GGGAAAGUCUUUCACUAAAUUAGCGCACCUCGACAGGCCGCAAGG-CUUGAAGAGAAAC-GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUA .((((....)))).........(((((((((..((((((....))-))))..)))....-.........((((.(((.((....)).)))...))))..)))))).(((((....))))) ( -42.20) >consensus CGAAAAGUGUUCGACUAAAUUAGCGCGCCUCGACAGACAGAACGG__UUGAAGAGAAAC_GGUGAAGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUA ......................(((((((((..((((.........))))..)))..............((((.((((((.....))).))).))))..)))))).(((((....))))) (-25.93 = -26.13 + 0.20)

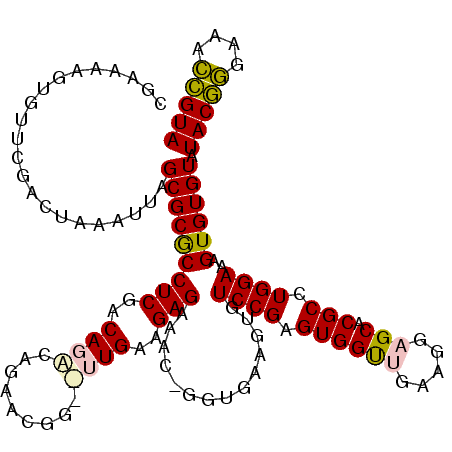
| Location | 3,206,391 – 3,206,508 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -36.21 |
| Energy contribution | -34.77 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1711 3206391 117 + 6264404 GGCGC---UGUCGAGAACCUGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAGAUCGCAAGA .((((---(((((.((((((....)))).)))))(((((.((.((.((((((.(....)))))))((((((....)))))).(((((.......)))))))))))))))))..))).... ( -45.80) >pau.1282 4174657 117 - 6537648 GGCGC---UGUCGAGAACCUGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAGAUCGCAAGA .((((---(((((.((((((....)))).)))))(((((.((.((.((((((.(....)))))))((((((....)))))).(((((.......)))))))))))))))))..))).... ( -45.80) >pel.854 1616217 113 + 5888780 AAUAG-CUUGAAGAGAAAC-GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUAUCAAGGGUUCAAAUCCCUUCUUCACCGCCACAUUC----- .....-............(-(((((((..((((.(((.((....)).)))...)))).....((.((((((....))))))))((((.......)))).))))))))........----- ( -35.80) >pf5.1100 2042069 114 + 7074893 AAUCGGUUUGAAGAGAUAC-GGUGAAGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAGGAAACUGUAUCGAGAGUUCGAAUCUCUCCUUCACCGCCACAUUC----- ..................(-(((((((..((((.(((((((...)))).))).))))......(.((((((....)))))))(((((.......)))))))))))))........----- ( -37.30) >pfo.993 1938154 114 + 6438405 AACAUGUUUGAAGAGAUAC-GGUGAAGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAGGAAACUGUAUCGAGAGUUCGAAUCUCUCCUUCACCGCCAAAUUC----- ..................(-(((((((..((((.(((((((...)))).))).))))......(.((((((....)))))))(((((.......)))))))))))))........----- ( -37.30) >ppk.1052 2069042 113 + 6181863 CAAGG-CUUGAAGAGAAAC-GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUAUCAAGGGUUCAAAUCCCUUCUUCACCGCCACAUUC----- ...((-(.....(.....)-(((((((..((((.(((.((....)).)))...)))).....((.((((((....))))))))((((.......)))).))))))))))......----- ( -37.40) >consensus AACGG__UUGAAGAGAAAC_GGUGAAGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUAUCGAGGGUUCGAAUCCCUCCUUCACCGCCACAUUC_____ ....................(((((((..((((.((((((.....))).))).))))......(.((((((....)))))))(((((.......)))))))))))).............. (-36.21 = -34.77 + -1.44)

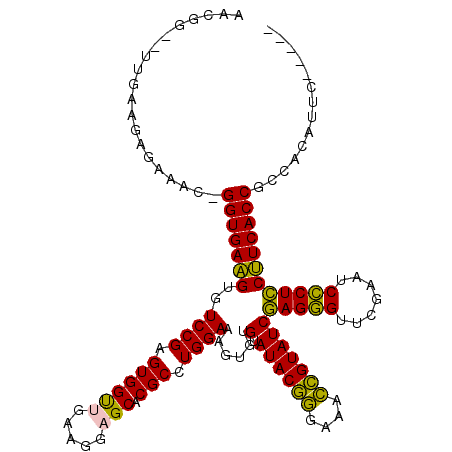

| Location | 3,206,391 – 3,206,508 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -28.32 |
| Energy contribution | -26.10 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1711 3206391 117 - 6264404 UCUUGCGAUCUGGCGGUGAGGGAGGGAUUCGAACCCUCGAUACGGUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACCUCACCAGGUUCUCGACA---GCGCC ...........((((((((((((((((.((((....))))(((((....)))))..(((.((.....)).))).))))))..((....))...)))))))..(((......)---))))) ( -36.80) >pau.1282 4174657 117 + 6537648 UCUUGCGAUCUGGCGGUGAGGGAGGGAUUCGAACCCUCGAUACGGUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACCUCACCAGGUUCUCGACA---GCGCC ...........((((((((((((((((.((((....))))(((((....)))))..(((.((.....)).))).))))))..((....))...)))))))..(((......)---))))) ( -36.80) >pel.854 1616217 113 - 5888780 -----GAAUGUGGCGGUGAAGAAGGGAUUUGAACCCUUGAUACGAUUUCUCGUAUACACACUUUCCAGGCGUGCUCCUUCGACCACUCGGACACUUCACC-GUUUCUCUUCAAG-CUAUU -----(((.(..((((((((((((((.......))))).((((((....))))))........(((.((.(((.((....)).))))))))..)))))))-))..)..)))...-..... ( -32.20) >pf5.1100 2042069 114 - 7074893 -----GAAUGUGGCGGUGAAGGAGAGAUUCGAACUCUCGAUACAGUUUCCUGUAUACACACUUUCCAGGCGUGCUCCUUAAGCCACUCGGACACUUCACC-GUAUCUCUUCAAACCGAUU -----(((....((((((((((((((.......))))).((((((....))))))........(((..(.(.(((.....)))).)..)))..)))))))-)).....)))......... ( -34.20) >pfo.993 1938154 114 - 6438405 -----GAAUUUGGCGGUGAAGGAGAGAUUCGAACUCUCGAUACAGUUUCCUGUAUACACACUUUCCAGGCGUGCUCCUUAAGCCACUCGGACACUUCACC-GUAUCUCUUCAAACAUGUU -----(((....((((((((((((((.......))))).((((((....))))))........(((..(.(.(((.....)))).)..)))..)))))))-)).....)))......... ( -34.20) >ppk.1052 2069042 113 - 6181863 -----GAAUGUGGCGGUGAAGAAGGGAUUUGAACCCUUGAUACGAUUUCUCGUAUACACACUUUCCAGGCGUGCUCCUUCGACCACUCGGACACUUCACC-GUUUCUCUUCAAG-CCUUG -----(((.(..((((((((((((((.......))))).((((((....))))))........(((.((.(((.((....)).))))))))..)))))))-))..)..)))...-..... ( -32.20) >consensus _____GAAUGUGGCGGUGAAGGAGGGAUUCGAACCCUCGAUACGGUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACUUCACC_GUUUCUCUUCAA__CCGUU ..............((((((((((((.......))))).((((((....)))))).(((.((.....)).))).........((....))...))))))).................... (-28.32 = -26.10 + -2.22)
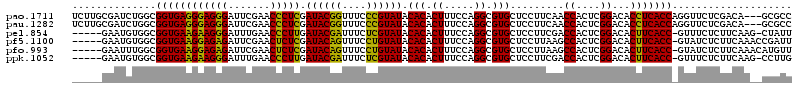
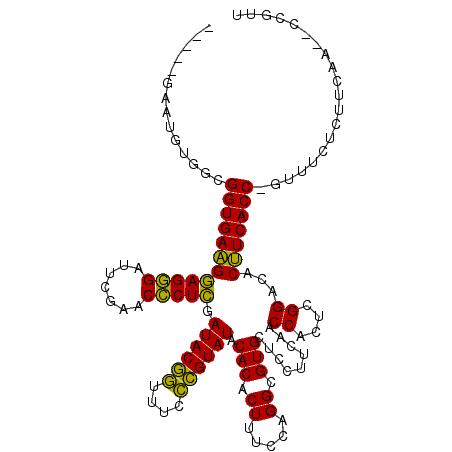
| Location | 3,206,407 – 3,206,527 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -35.51 |
| Energy contribution | -34.48 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1711 3206407 120 + 6264404 UGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAGAUCGCAAGAAGGCCCGGCUAAUCAAGUU .(((((((..((((.((((((.....))).))).))))......(.((((((....)))))))(((((.......))))))))))))(((...((....)).)))............... ( -45.70) >pau.1282 4174673 120 - 6537648 UGGUGAGGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGGGAAACCGUAUCGAGGGUUCGAAUCCCUCCCUCACCGCCAGAUCGCAAGAAGGCCCGGCUAAUCAAGUU .(((((((..((((.((((((.....))).))).))))......(.((((((....)))))))(((((.......))))))))))))(((...((....)).)))............... ( -45.70) >pel.854 1616235 95 + 5888780 -GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUAUCAAGGGUUCAAAUCCCUUCUUCACCGCCACAUUC------------------------ -(((((((..((((.(((.((....)).)))...)))).....((.((((((....))))))))((((.......)))).))))))).........------------------------ ( -34.20) >psp.1108 3785607 117 - 5928787 -GGUGAGGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAAGAAAUUGUAUCGAGGGUUCGAAUCCCUCCUUCACCGCCAUAUU--AAGAAAACCCUGUGAAUUCAGUU -(((((((..((((.(((((((...)))).))).))))......(.((((((....)))))))(((((.......))))))))))))........--.........(((......))).. ( -35.20) >pfo.993 1938173 95 + 6438405 -GGUGAAGUGUCCGAGUGGCUUAAGGAGCACGCCUGGAAAGUGUGUAUACAGGAAACUGUAUCGAGAGUUCGAAUCUCUCCUUCACCGCCAAAUUC------------------------ -(((((((..((((.(((((((...)))).))).))))......(.((((((....)))))))(((((.......)))))))))))).........------------------------ ( -35.70) >ppk.1052 2069060 95 + 6181863 -GGUGAAGUGUCCGAGUGGUCGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAAUCGUAUCAAGGGUUCAAAUCCCUUCUUCACCGCCACAUUC------------------------ -(((((((..((((.(((.((....)).)))...)))).....((.((((((....))))))))((((.......)))).))))))).........------------------------ ( -34.20) >consensus _GGUGAAGUGUCCGAGUGGUUGAAGGAGCACGCCUGGAAAGUGUGUAUACGAGAAACCGUAUCGAGGGUUCGAAUCCCUCCUUCACCGCCACAUUC________________________ .(((((((..((((.((((((.....))).))).))))......(.((((((....)))))))(((((.......))))))))))))................................. (-35.51 = -34.48 + -1.02)

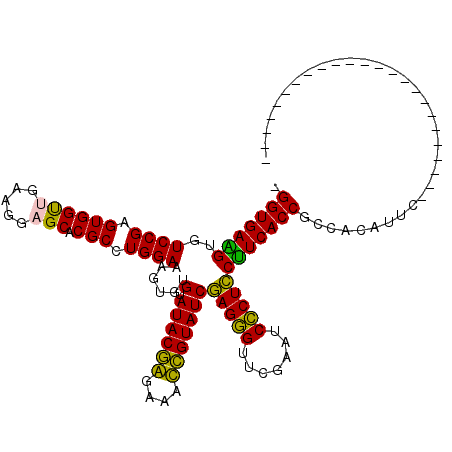
| Location | 3,206,407 – 3,206,527 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -27.46 |
| Energy contribution | -25.63 |
| Covariance contribution | -1.83 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1711 3206407 120 - 6264404 AACUUGAUUAGCCGGGCCUUCUUGCGAUCUGGCGGUGAGGGAGGGAUUCGAACCCUCGAUACGGUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACCUCACCA ..........(((((..(.......)..)))))(((((((((((((.((((....))))(((((....)))))..(((.((.....)).))).))))))..((....))...))))))). ( -42.40) >pau.1282 4174673 120 + 6537648 AACUUGAUUAGCCGGGCCUUCUUGCGAUCUGGCGGUGAGGGAGGGAUUCGAACCCUCGAUACGGUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACCUCACCA ..........(((((..(.......)..)))))(((((((((((((.((((....))))(((((....)))))..(((.((.....)).))).))))))..((....))...))))))). ( -42.40) >pel.854 1616235 95 - 5888780 ------------------------GAAUGUGGCGGUGAAGAAGGGAUUUGAACCCUUGAUACGAUUUCUCGUAUACACACUUUCCAGGCGUGCUCCUUCGACCACUCGGACACUUCACC- ------------------------(((.(((.(((((..((((((.............((((((....)))))).(((.((.....)).))).))))))...))).))..))))))...- ( -25.20) >psp.1108 3785607 117 + 5928787 AACUGAAUUCACAGGGUUUUCUU--AAUAUGGCGGUGAAGGAGGGAUUCGAACCCUCGAUACAAUUUCUUGUAUACACACUUUCCAGGCGUGCUCCUUAAGCCACUCGGACACCUCACC- ..(((......)))(((.(((..--....(((.((((...(((((.......))))).((((((....))))))...))))..)))(((...........)))....))).))).....- ( -27.90) >pfo.993 1938173 95 - 6438405 ------------------------GAAUUUGGCGGUGAAGGAGAGAUUCGAACUCUCGAUACAGUUUCCUGUAUACACACUUUCCAGGCGUGCUCCUUAAGCCACUCGGACACUUCACC- ------------------------.........((((((((((((.......))))).((((((....))))))........(((..(.(.(((.....)))).)..)))..)))))))- ( -27.80) >ppk.1052 2069060 95 - 6181863 ------------------------GAAUGUGGCGGUGAAGAAGGGAUUUGAACCCUUGAUACGAUUUCUCGUAUACACACUUUCCAGGCGUGCUCCUUCGACCACUCGGACACUUCACC- ------------------------(((.(((.(((((..((((((.............((((((....)))))).(((.((.....)).))).))))))...))).))..))))))...- ( -25.20) >consensus ________________________GAAUCUGGCGGUGAAGGAGGGAUUCGAACCCUCGAUACGAUUUCCCGUAUACACACUUUCCAGGCGUGCUCCUUCAACCACUCGGACACCUCACC_ .................................((((((((((((.......))))).((((((....)))))).(((.((.....)).))).........((....))...))))))). (-27.46 = -25.63 + -1.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:24 2007