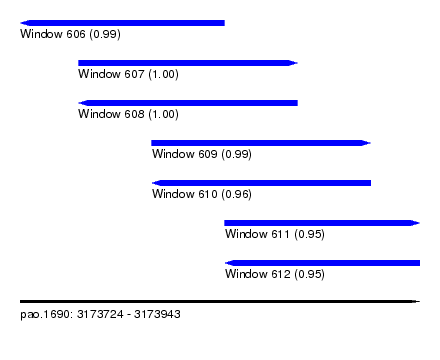
| Sequence ID | pao.1690 |
|---|---|
| Location | 3,173,724 – 3,173,943 |
| Length | 219 |
| Max. P | 0.999419 |

| Location | 3,173,724 – 3,173,836 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -49.27 |
| Consensus MFE | -44.28 |
| Energy contribution | -44.15 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173724 112 - 6264404 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAUUCUA-CAGCCUGGCUU--CGGUGGGGUUG-----GUGAAA .(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))......(((..-((((((.((..--..)).))))))-----..))). ( -49.70) >pau.1303 4141991 112 + 6537648 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAUUCUA-CAGCCUGGCUU--CGGCGGGGUUG-----GUGAAA .(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))......(((..-((((((.((..--..)).))))))-----..))). ( -51.20) >pel.884 4228285 117 + 5888780 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGUUUCUC-CGGCACUGCUU--CGGCAGGGCAGGAAAAGAAAAU .(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).....((((((-(.((.((((..--..)))).)).)))...)))).. ( -49.70) >ppk.1076 4079031 119 + 6181863 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGUUUCUC-CGGCACUGCUUAGCGGCGGGGCAGGAAAAGAAAAC .(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).....((((((-(.((.(((((....))))).)).)))...)))).. ( -46.40) >pfo.2462 4729381 116 - 6438405 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAUUUUAAACAGCAACGCUUUCGGGCGGUGCUG----AGUGAAA .(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).....(((((..(((((.((((....)))).)))))----..))))) ( -48.10) >pst.1067 4440837 116 + 6397126 UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAUUUUUAAGCAAUCUUGCUCACGGGCAAGGUUG----AGUGAAA .(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).....(((((..((((((((((....))))))))))----..))))) ( -50.50) >consensus UGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAUUUCUA_CAGCAUUGCUU__CGGCGGGGCUG____AGUGAAA .(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))............((((((((((....))))))))))........... (-44.28 = -44.15 + -0.13)

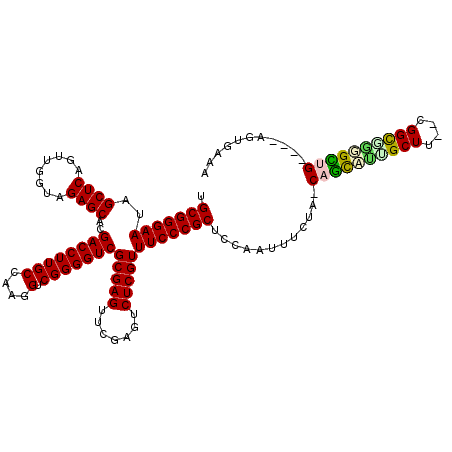
| Location | 3,173,756 – 3,173,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -45.75 |
| Consensus MFE | -45.35 |
| Energy contribution | -45.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173756 120 + 6264404 AUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).((((((.....((((((.......))))))....)))))) ( -46.20) >pau.1303 4142023 120 - 6537648 AUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).((((((.....((((((.......))))))....)))))) ( -46.20) >pel.884 4228322 120 - 5888780 AACUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))...((((.....((((((.......))))))....)))).. ( -45.30) >ppk.1076 4079070 120 - 6181863 AACUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))...((((.....((((((.......))))))....)))).. ( -45.30) >pfo.2462 4729417 120 + 6438405 AAUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))...((((.....((((((.......))))))....)))).. ( -45.30) >pst.1067 4440873 120 - 6397126 AAAUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).((((((.....((((((.......))))))....)))))) ( -46.20) >consensus AAUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGU .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))..(((((.....((((((.......))))))....))))). (-45.35 = -45.35 + 0.00)
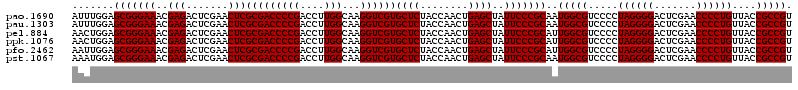
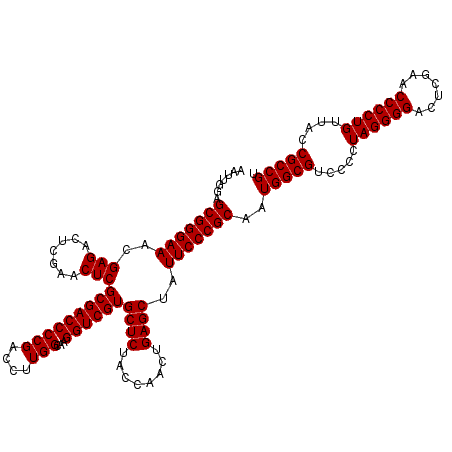
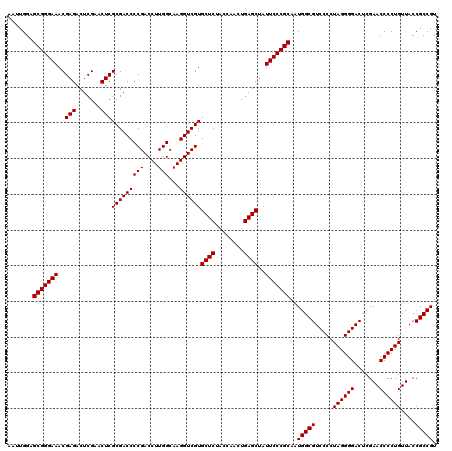
| Location | 3,173,756 – 3,173,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -50.50 |
| Energy contribution | -50.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173756 120 - 6264404 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAU ..(((((.(((.(((..(((((((((....)))))....((((((.....((((........)))).((((((.....))))))...))))))..))))..))))))..)))))...... ( -50.60) >pau.1303 4142023 120 + 6537648 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAU ..(((((.(((.(((..(((((((((....)))))....((((((.....((((........)))).((((((.....))))))...))))))..))))..))))))..)))))...... ( -50.60) >pel.884 4228322 120 + 5888780 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGUU ..(((((.(((.(((..(((((((((....)))))((((((..(((.....))).))))))....(.(((((((((....).)))))))).)...))))..))))))..)))))...... ( -50.60) >ppk.1076 4079070 120 + 6181863 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGUU ..(((((.(((.(((..(((((((((....)))))((((((..(((.....))).))))))....(.(((((((((....).)))))))).)...))))..))))))..)))))...... ( -50.60) >pfo.2462 4729417 120 - 6438405 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAUU ..(((((.(((.(((..(((((((((....)))))((((((..(((.....))).))))))....(.(((((((((....).)))))))).)...))))..))))))..)))))...... ( -50.60) >pst.1067 4440873 120 + 6397126 ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAUUU ..(((((.(((.(((..(((((((((....)))))....((((((.....((((........)))).((((((.....))))))...))))))..))))..))))))..)))))...... ( -50.60) >consensus ACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAUU ..((((....((((((.......))))).)....))))...(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))....... (-50.50 = -50.50 + 0.00)

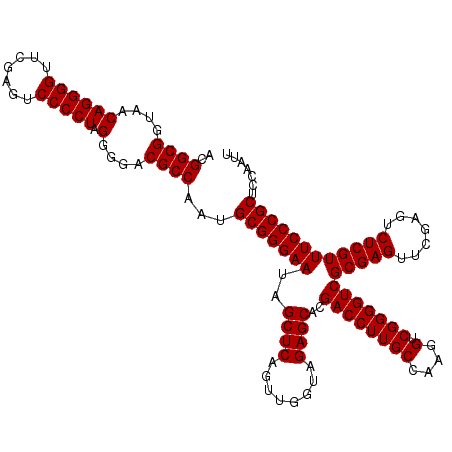
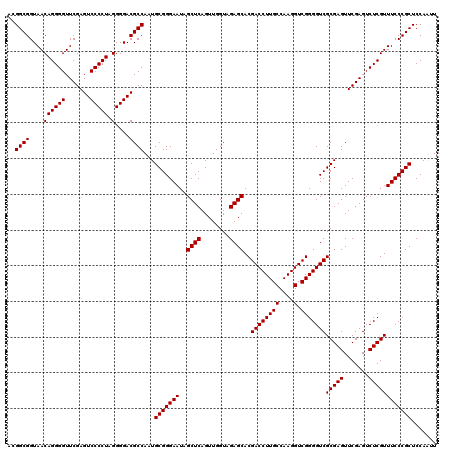
| Location | 3,173,796 – 3,173,916 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -48.53 |
| Consensus MFE | -46.70 |
| Energy contribution | -46.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173796 120 + 6264404 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCG ....((..((....((((........))))....)).))....((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))). ( -52.20) >pau.1303 4142063 120 - 6537648 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCG ....((..((....((((........))))....)).))....((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))). ( -52.20) >pel.884 4228362 119 - 5888780 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC- ....((..((....((((........))))....)).))......((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..- ( -46.70) >ppk.1076 4079110 119 - 6181863 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC- ....((..((....((((........))))....)).))......((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..- ( -46.70) >pfo.2462 4729457 119 + 6438405 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC- ....((..((....((((........))))....)).))......((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..- ( -46.70) >pst.1067 4440913 119 - 6397126 CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC- ....((..((....((((........))))....)).))......((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..- ( -46.70) >consensus CUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC_ ....((..((....((((........))))....)).))......((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))... (-46.70 = -46.70 + -0.00)

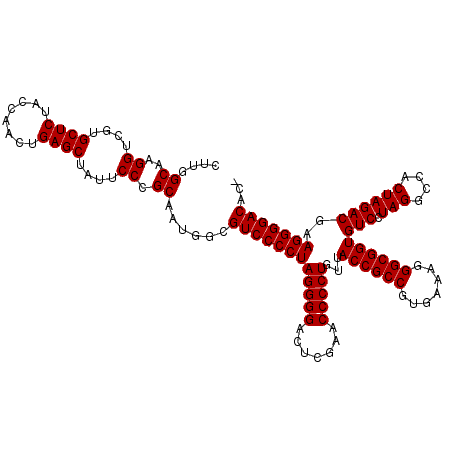
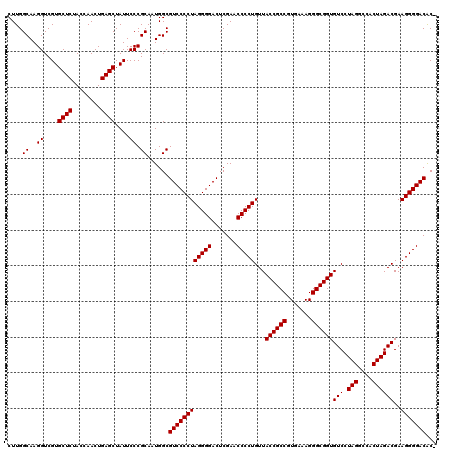
| Location | 3,173,796 – 3,173,916 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -47.83 |
| Consensus MFE | -48.06 |
| Energy contribution | -47.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.04 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173796 120 - 6264404 CGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG .(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -49.30) >pau.1303 4142063 120 + 6537648 CGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG .(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -49.30) >pel.884 4228362 119 + 5888780 -GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG -(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -47.10) >ppk.1076 4079110 119 + 6181863 -GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG -(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -47.10) >pfo.2462 4729457 119 - 6438405 -GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG -(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -47.10) >pst.1067 4440913 119 + 6397126 -GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG -(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... ( -47.10) >consensus _GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAG .(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))....(((((....((((........)))).....))))).... (-48.06 = -47.83 + -0.22)

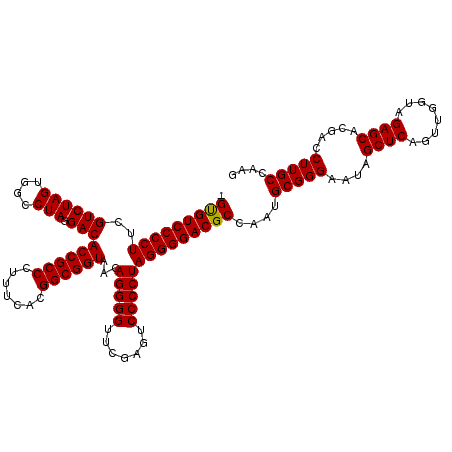
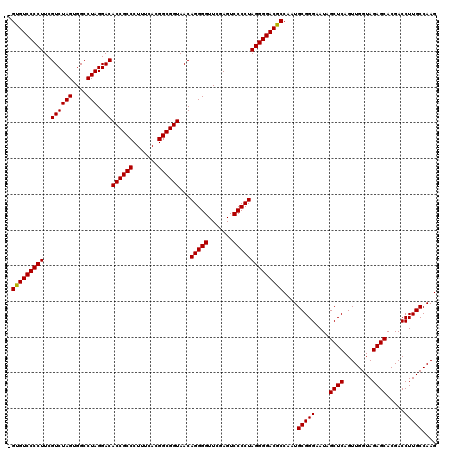
| Location | 3,173,836 – 3,173,943 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -42.05 |
| Consensus MFE | -37.20 |
| Energy contribution | -37.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173836 107 + 6264404 AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCGGCCCGGAACUUA-------------CAACAGAUCUUUCCG ...((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))))....(((((....-------------..........))))) ( -45.34) >pau.1303 4142103 107 - 6537648 AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCGGCCCGGAACUUA-------------CAACAGAUCUUUCCG ...((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))))....(((((....-------------..........))))) ( -45.34) >pel.884 4228402 94 - 5888780 UUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC--------GC-UA-------------CAACA----UUCACU .((((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..--------))-))-------------.....----...... ( -39.10) >ppk.1076 4079150 107 - 6181863 UUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC--------GC-UGCUGGCAUUACUGCCUGCU----UUCACU ..(((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..--------))-)((.((((....)))).)).----...... ( -46.00) >pf5.1056 5107985 94 - 7074893 AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC--------GC-UA-------------CAGCA----UUCACU .((((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))..--------))-))-------------.....----...... ( -39.20) >pfo.2462 4729497 94 + 6438405 UUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC--------AC-UA-------------CAACA----UUCACU ...(.((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))).)--------..-..-------------.....----...... ( -37.30) >consensus AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAC________AC_UA_____________CAACA____UUCACU .....((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))........................................... (-37.20 = -37.20 + -0.00)

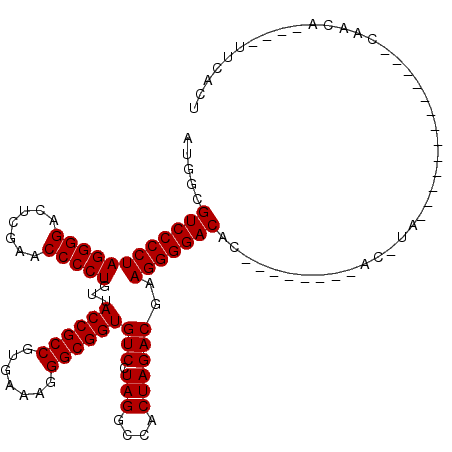
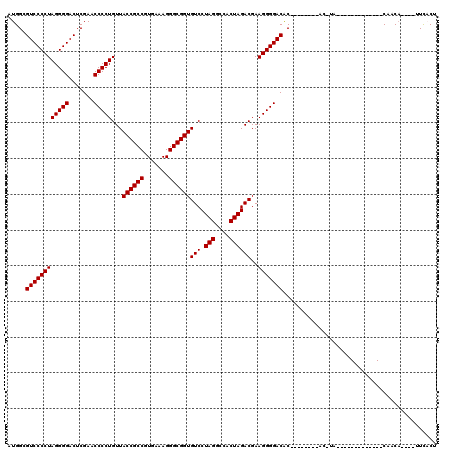
| Location | 3,173,836 – 3,173,943 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -43.18 |
| Consensus MFE | -38.96 |
| Energy contribution | -38.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1690 3173836 107 - 6264404 CGGAAAGAUCUGUUG-------------UAAGUUCCGGGCCGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAU ((((....((((((.-------------...((((..((((((..............))))))..))))((((((.......))))))))))))..)))).(((((....)))))..... ( -46.84) >pau.1303 4142103 107 + 6537648 CGGAAAGAUCUGUUG-------------UAAGUUCCGGGCCGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAU ((((....((((((.-------------...((((..((((((..............))))))..))))((((((.......))))))))))))..)))).(((((....)))))..... ( -46.84) >pel.884 4228402 94 + 5888780 AGUGAA----UGUUG-------------UA-GC--------GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAA ......----.....-------------..-(.--------(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))).. ( -38.60) >ppk.1076 4079150 107 + 6181863 AGUGAA----AGCAGGCAGUAAUGCCAGCA-GC--------GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAA ......----.((.((((....)))).)).-(.--------(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))).. ( -48.60) >pf5.1056 5107985 94 + 7074893 AGUGAA----UGCUG-------------UA-GC--------GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAU .(((..----.((..-------------..-))--------(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))))) ( -39.60) >pfo.2462 4729497 94 - 6438405 AGUGAA----UGUUG-------------UA-GU--------GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAA ......----.....-------------..-(.--------(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))).. ( -38.60) >consensus AGUGAA____UGUUG_____________UA_GC________GUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAA .........................................(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))... (-38.96 = -38.73 + -0.22)

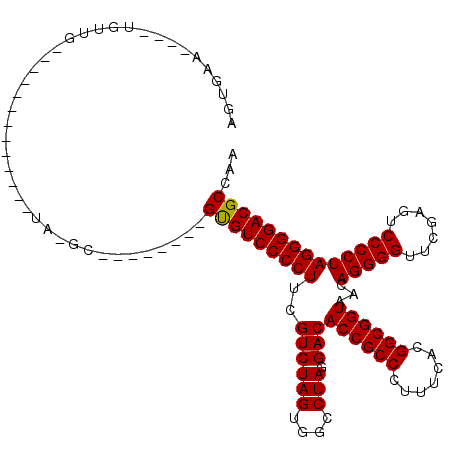
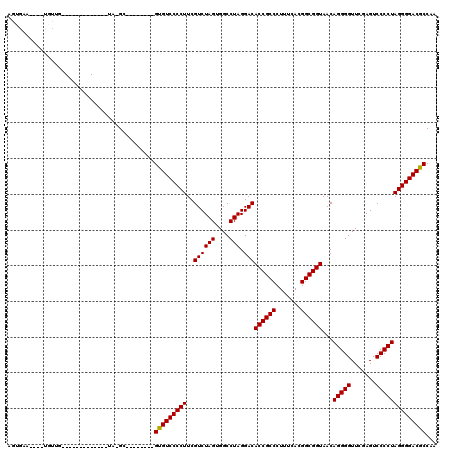
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:19 2007