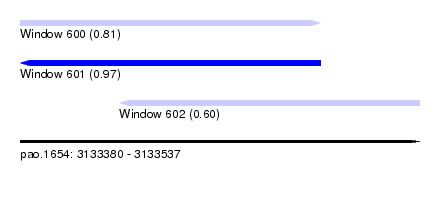
| Sequence ID | pao.1654 |
|---|---|
| Location | 3,133,380 – 3,133,537 |
| Length | 157 |
| Max. P | 0.971980 |

| Location | 3,133,380 – 3,133,498 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -29.76 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1654 3133380 118 + 6264404 GGGCGCAAACGAAGAAAGGCGACCCUUUCGGAUCGCCU-UUCUUGCCCCGCCAGCAGCGCGGGAUUU-GUUUGGUAGGCACGAUUGGAUUCGAACCAACGACCCCCACCAUGUCAAGGUG ((((((((((((((((((((((.((....)).))))))-)))....((((((....).))))).)))-)))))...((..(((......)))..))..)).))).((((.......)))) ( -47.30) >pau.1345 4096759 118 - 6537648 GGGCGCAAACGAAAAAAGGCGACCCUUUCGGAUCGCCU-UUCUUGCCCCGCCAGCAGUGCGGGAUUU-GUUUGGUAGGCACGAUUGGAUUCGAACCAACGACCCCCACCAUGUCAAGGUG (((((((((((((.((((((((.((....)).))))))-)).....((((((....).))))).)))-)))))...((..(((......)))..))..)).))).((((.......)))) ( -43.40) >psp.1547 2950246 113 - 5928787 ------AACAAA-AAAGGGCCAGCCUUUCGGCUGACCCUUUUCUUACCGCCCAGCAGAGCGGAUUUUUAUCUGGUAGGCGCGAUUGGACUCGAACCAACGACCCCCACCAUGUCAAGGUG ------.....(-((((((.(((((....))))).)))))))....((((.(....).))))(((((.((.((((.((.(((.((((.......)))))).).)).)))).)).))))). ( -37.10) >pf5.1252 4765091 112 - 7074893 ------ACAAAA-AAGGGGCUCACCUUUCGGUGAGCCCCUUCUAGACCGCCCAGCAGAGCGGAUUUU-GUUUGGUAGGCGCGAUUGGACUCGAACCAACGACCCCCACCAUGUCAAGGUG ------......-((((((((((((....)))))))))))).....((((.(....).))))(((((-(..((((.((.(((.((((.......)))))).).)).))))...)))))). ( -46.10) >pfo.1100 4253967 111 - 6438405 ------AACAAA-AAAGGGCCCACCUUUCGGUGAGCCC-UUCUAGACCGCCCAGCAGAGCGGAUUUU-GUUUGGUAGGCGCGAUUGGACUCGAACCAACGACCCCCACCAUGUCAAGGUG ------......-.((((((.((((....)))).))))-)).....((((.(....).))))(((((-(..((((.((.(((.((((.......)))))).).)).))))...)))))). ( -38.00) >pss.1485 3284444 112 - 6093698 ------AAACGA-AAAGGGCCAGCCUCUCGGCUGACCCUUUUCUUACCGCCCAGCAGAGCGGAUUUU-GUCUGGUAGGCGCGAUUGGACUCGAACCAACGACCCCCACCAUGUCAAGGUG ------....((-((((((.(((((....))))).))))))))...((((.(....).))))(((((-(.(((((.((.(((.((((.......)))))).).)).)))).).)))))). ( -44.20) >consensus ______AAAAAA_AAAGGGCCAACCUUUCGGAUAGCCC_UUCUUGACCGCCCAGCAGAGCGGAUUUU_GUUUGGUAGGCGCGAUUGGACUCGAACCAACGACCCCCACCAUGUCAAGGUG ................(((((((((....)))))))))........((((.(....).))))(((((.(..((((.((..((.((((.......))))))...)).))))..).))))). (-29.76 = -29.68 + -0.08)

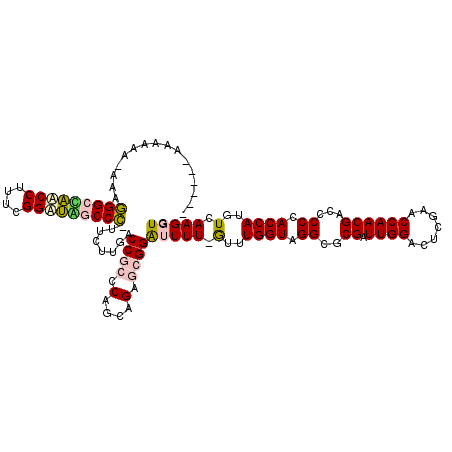
| Location | 3,133,380 – 3,133,498 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -58.15 |
| Consensus MFE | -48.62 |
| Energy contribution | -46.98 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.32 |
| Mean z-score | -5.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1654 3133380 118 - 6264404 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAAUCCAAUCGUGCCUACCAAAC-AAAUCCCGCGCUGCUGGCGGGGCAAGAA-AGGCGAUCCGAAAGGGUCGCCUUUCUUCGUUUGCGCCC ((((.......)))).((((.((.(((.(((......))).))).))(((((-....(((((.(....))))))(.(((((-(((((((((....)))))))))))))))))))).)))) ( -56.80) >pau.1345 4096759 118 + 6537648 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAAUCCAAUCGUGCCUACCAAAC-AAAUCCCGCACUGCUGGCGGGGCAAGAA-AGGCGAUCCGAAAGGGUCGCCUUUUUUCGUUUGCGCCC ((((.......)))).((((.((.(((.(((......))).))).))(((((-....(((((.(....))))))(.(((((-(((((((((....)))))))))))))))))))).)))) ( -54.20) >psp.1547 2950246 113 + 5928787 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAGAUAAAAAUCCGCUCUGCUGGGCGGUAAGAAAAGGGUCAGCCGAAAGGCUGGCCCUUU-UUUGUU------ ..........(((((((.((.(((((.......))))).)).))))))).........((((((....))))))..(((((((((((((((....))))))))))))-)))...------ ( -58.50) >pf5.1252 4765091 112 + 7074893 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAAAC-AAAAUCCGCUCUGCUGGGCGGUCUAGAAGGGGCUCACCGAAAGGUGAGCCCCUU-UUUUGU------ ..........(((((((.((.(((((.......))))).)).))))))).((-(((..((((((....))))))....(((((((((((((....))))))))))))-))))))------ ( -61.20) >pfo.1100 4253967 111 + 6438405 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAAAC-AAAAUCCGCUCUGCUGGGCGGUCUAGAA-GGGCUCACCGAAAGGUGGGCCCUUU-UUUGUU------ ..........(((((((.((.(((((.......))))).)).)))))))(((-(((..((((((....))))))....(((-(((((((((....))))))))))))-))))))------ ( -57.90) >pss.1485 3284444 112 + 6093698 CACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAGAC-AAAAUCCGCUCUGCUGGGCGGUAAGAAAAGGGUCAGCCGAGAGGCUGGCCCUUU-UCGUUU------ ....(((...(((((((.((.(((((.......))))).)).)))))))..)-))...((((((....))))))...((((((((((((((....))))))))))))-))....------ ( -60.30) >consensus CACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAAAC_AAAAUCCGCUCUGCUGGGCGGUCAAGAA_GGGCCAACCGAAAGGCUGGCCCUUU_UCGUUU______ ..........(((((((.(..(((((.......)))))..).))))))).........((((((....))))))........(((((((((....)))))))))................ (-48.62 = -46.98 + -1.63)

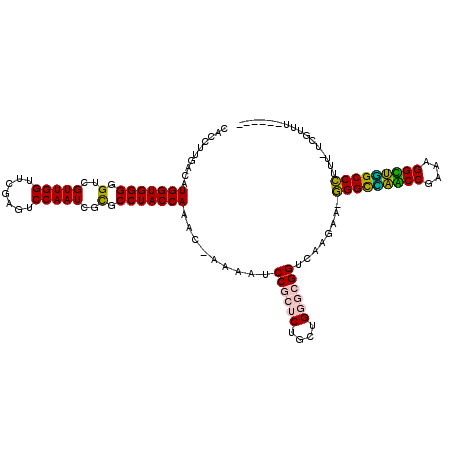
| Location | 3,133,419 – 3,133,537 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
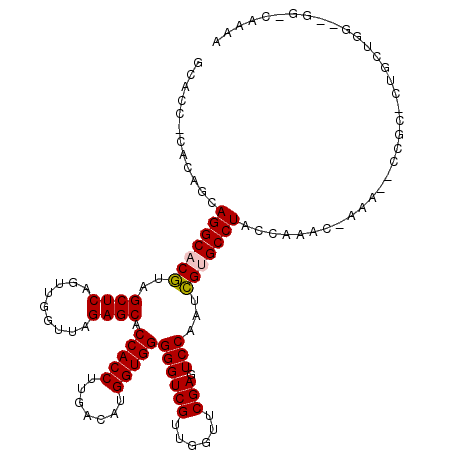
>pao.1654 3133419 118 - 6264404 GCGCC-AUCAGCAGGCACGUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAAUCCAAUCGUGCCUACCAAAC-AAAUCCCGCGCUGCUGGCGGGGCAAGA ((.((-..((((((((.....((((.........)))).(((((.......)))))(((...(((((.(((......))).....)))))..-....))))).))))))..)).)).... ( -46.50) >pau.1345 4096798 118 + 6537648 GCGCC-AUCAGCAGGCACGUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAAUCCAAUCGUGCCUACCAAAC-AAAUCCCGCACUGCUGGCGGGGCAAGA ((.((-..((((((((.....((((.........)))).(((((.......)))))(((...(((((.(((......))).....)))))..-....))))).))))))..)).)).... ( -46.50) >psp.1547 2950279 119 + 5928787 GGACC-CACAGCAGGCACGUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAGAUAAAAAUCCGCUCUGCUGGGCGGUAAGAA ..(((-(.((((((((.....((((.........))))............(((((((.((.(((((.......))))).)).)))))))...........)).)))))).).)))..... ( -42.60) >pel.1234 3612857 97 + 5888780 GCACCACACGACAGGCACAUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUUGUGCCUACCAAAU-----------------------CAGAA ............(((((((..((((.........)))).(((((.......)))))((((((......))).)))...))))))).......-----------------------..... ( -29.70) >pss.1485 3284477 118 + 6093698 CGACC-CACAGCAGGCACGUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGCGCCUACCAGAC-AAAAUCCGCUCUGCUGGGCGGUAAGAA ..(((-(.((((((((.....((((.........))))......(((...(((((((.((.(((((.......))))).)).)))))))..)-)).....)).)))))).).)))..... ( -43.50) >ppk.1430 3385042 97 + 6181863 GCACCACACGACAGGCACAUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUUGUGCCUACCAAAU-----------------------CGAAA ............(((((((..((((.........)))).(((((.......)))))((((((......))).)))...))))))).......-----------------------..... ( -29.70) >consensus GCACC_CACAGCAGGCACGUAGCUCAGUUGGUUAGAGCACCACCUUGACAUGGUGGGGGUCGUUGGUUCGAGUCCAAUCGUGCCUACCAAAC_AAA__CCGC_CUGCUGG__GG_CAAAA ............(((((((..((((.........)))).(((((.......)))))((((((......))).)))...)))))))................................... (-28.84 = -28.73 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:14:08 2007