
| Sequence ID | pao.1825 |
|---|---|
| Location | 3,414,291 – 3,414,400 |
| Length | 109 |
| Max. P | 0.989878 |

| Location | 3,414,291 – 3,414,398 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.92 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -33.15 |
| Energy contribution | -30.93 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
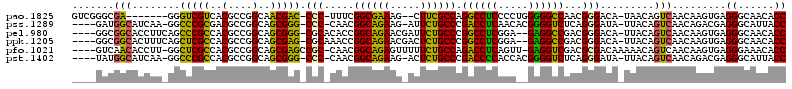
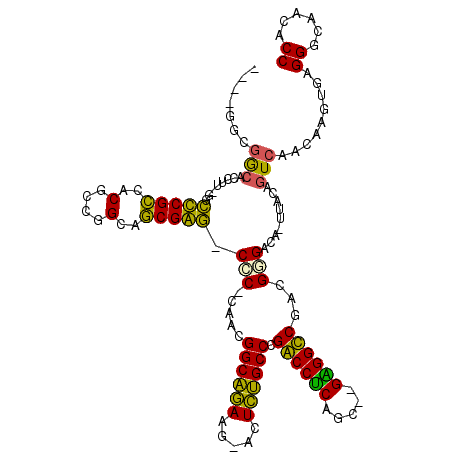
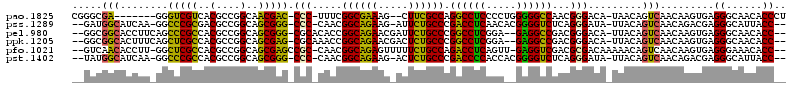
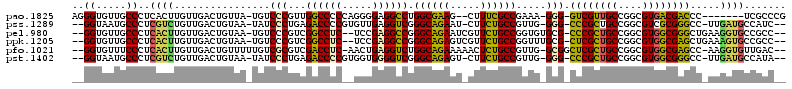
>pao.1825 3414291 107 + 6264404 GUCGGGCGA-------GGGUCGUCACGCCGGCAACGAC-CCC-UUUCGGCGAAAG--CUUCGCCAGGCCUCCCCUGGGGGCCAACGGGACA-UAACAGUCAACAAGUGAGGGCAACACC (((.(((((-------(((((((..(....)..)))))-)).-....(((....)--))))))).(((((((...))))))).....))).-.....(((..(....)..)))...... ( -44.10) >pss.1289 3634006 110 - 6093698 ----GAUGGCAUCAA-GGCCCGCGACGCCGGCAGCGGG-CCC-CAACGGCAGAAG-AUUCUGCCCGACCUCAACACGGGGUCUCAGGGAUA-UUACAGUCAACAGACGAGGGCAUUACC ----((((.(.((..-(((((((..(....)..)))))-))(-(...((((((..-..)))))).((((((.....))))))...))....-.....(((....))))).).))))... ( -44.20) >pel.980 4056794 111 - 5888780 ----GGCGGCACCUUCAGCCCGCCACGCCGGCAGCGGG-CGCACACCGGCAGAACGAUUCUGCCCGGCCUCGGA--GAGGCCGACGGGACA-UUACAGUCAACAAGUGAGGGCAACACC ----.(..((.(((...((((((..(....)..)))))-).(((.((((((((.....))))))(((((((...--)))))))..))(((.-.....))).....))))))))..)... ( -48.30) >ppk.1205 3793131 111 - 6181863 ----GGCGGCACUUUCAGCUCGCCACGCCGGCAGCGAG-CGCAAACCGGCAGAACGACUCUGCCCGGCCUCGGA--GAGGCCGACGGGACA-UUACAGUCAACAAGUGAGGGCAACACC ----((((((.......)).))))..(((((..((...-.))...)))))..........(((((((((((...--))))))(((.(....-...).))).........)))))..... ( -44.50) >pfo.1021 4449300 112 - 6438405 ----GUCAACACCUU-GGCUCGCCACGCCGGCAGCGAGCCGC-CAACGGCAGAGUUUUUCUGCCAGACCUCAGUU-GAGGUCGACGCGACAAAAACAGUCAACAAGUGAGGGAAACACC ----.......((((-(((((((..(....)..)))))))((-....(((((((...))))))).((((((....-))))))...))(((.......))).......))))........ ( -41.90) >pst.1402 3839454 110 - 6397126 ----UAUGGCAUCAA-GGCCCGCCACGCCGGCAGCGGG-CCC-CAACGGCAGAAG-ACUCUGCCCGACCCCACCACGGGGUCUCAGGGAUA-UUACAGUCAACAGACGAGGGCAUUACC ----...........-.(((((((.(((.....)))))-)((-(...((((((..-..)))))).((((((.....))))))...)))...-.....(((....)))..))))...... ( -46.00) >consensus ____GGCGGCACCUU_GGCCCGCCACGCCGGCAGCGAG_CCC_CAACGGCAGAAG_ACUCUGCCCGACCUCAGC__GAGGCCGACGGGACA_UUACAGUCAACAAGUGAGGGCAACACC .......(((........(((((..(....)..))))).(((.....((((((.....)))))).((((((.....))))))...))).........))).........((......)) (-33.15 = -30.93 + -2.22)
| Location | 3,414,291 – 3,414,398 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.92 |
| Mean single sequence MFE | -48.52 |
| Consensus MFE | -34.25 |
| Energy contribution | -32.53 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
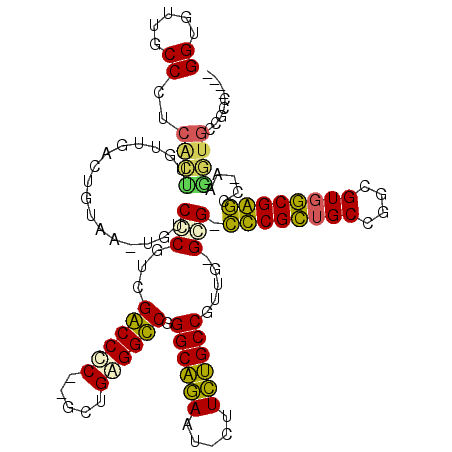
>pao.1825 3414291 107 - 6264404 GGUGUUGCCCUCACUUGUUGACUGUUA-UGUCCCGUUGGCCCCCAGGGGAGGCCUGGCGAAG--CUUUCGCCGAAA-GGG-GUCGUUGCCGGCGUGACGACCC-------UCGCCCGAC ((((......((.......(((.....-.))).....((((((....)).)))).(((((..--...)))))))..-(((-(((((..(....)..)))))))-------))))).... ( -42.10) >pss.1289 3634006 110 + 6093698 GGUAAUGCCCUCGUCUGUUGACUGUAA-UAUCCCUGAGACCCCGUGUUGAGGUCGGGCAGAAU-CUUCUGCCGUUG-GGG-CCCGCUGCCGGCGUCGCGGGCC-UUGAUGCCAUC---- (((..(((((..(((....))).....-.........((((.(.....).)))))))))..))-)...((.(((..-(((-(((((.((....)).)))))))-)..))).))..---- ( -44.60) >pel.980 4056794 111 + 5888780 GGUGUUGCCCUCACUUGUUGACUGUAA-UGUCCCGUCGGCCUC--UCCGAGGCCGGGCAGAAUCGUUCUGCCGGUGUGCG-CCCGCUGCCGGCGUGGCGGGCUGAAGGUGCCGCC---- ((((..(((.(((......(((.....-.)))((..(((((((--...)))))))((((((.....)))))))).....(-(((((..(....)..))))))))).)))..))))---- ( -54.00) >ppk.1205 3793131 111 + 6181863 GGUGUUGCCCUCACUUGUUGACUGUAA-UGUCCCGUCGGCCUC--UCCGAGGCCGGGCAGAGUCGUUCUGCCGGUUUGCG-CUCGCUGCCGGCGUGGCGAGCUGAAAGUGCCGCC---- ((((..((..(((...((.((((....-........(((((((--...)))))))(((((((...))))))))))).))(-(((((..(....)..)))))))))..))..))))---- ( -48.40) >pfo.1021 4449300 112 + 6438405 GGUGUUUCCCUCACUUGUUGACUGUUUUUGUCGCGUCGACCUC-AACUGAGGUCUGGCAGAAAAACUCUGCCGUUG-GCGGCUCGCUGCCGGCGUGGCGAGCC-AAGGUGUUGAC---- ........(..(((((((..((...............((((((-....)))))).((((((.....))))))))..-))(((((((..(....)..)))))))-.)))))..)..---- ( -49.00) >pst.1402 3839454 110 + 6397126 GGUAAUGCCCUCGUCUGUUGACUGUAA-UAUCCCUGAGACCCCGUGGUGGGGUCGGGCAGAGU-CUUCUGCCGUUG-GGG-CCCGCUGCCGGCGUGGCGGGCC-UUGAUGCCAUA---- (((..(((((..(((....))).....-.........((((((.....)))))))))))....-........((..-(((-(((((..(....)..)))))))-)..)))))...---- ( -53.00) >consensus GGUGUUGCCCUCACUUGUUGACUGUAA_UGUCCCGUCGACCCC__GCUGAGGCCGGGCAGAAU_CUUCUGCCGUUG_GCG_CCCGCUGCCGGCGUGGCGAGCC_AAGGUGCCGCC____ ((.....))..((((................(((...((((((.....)))))).((((((.....)))))).....))).((((((((....)))))))).....))))......... (-34.25 = -32.53 + -1.72)

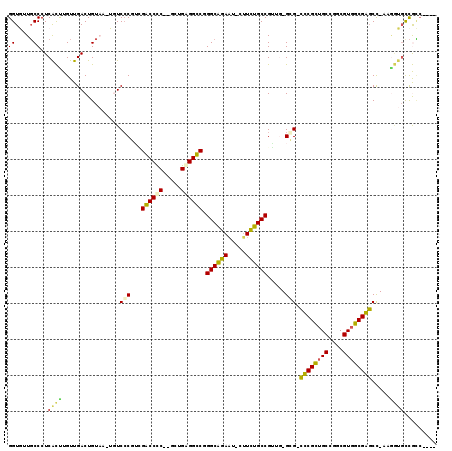
| Location | 3,414,293 – 3,414,400 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.92 |
| Mean single sequence MFE | -45.15 |
| Consensus MFE | -33.28 |
| Energy contribution | -31.07 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1825 3414293 107 + 6264404 CGGGCGA-------GGGUCGUCACGCCGGCAACGAC-CCC-UUUCGGCGAAAG--CUUCGCCAGGCCUCCCCUGGGGGCCAACGGGACA-UAACAGUCAACAAGUGAGGGCAACACCCU (((((((-------(((((((..(....)..)))))-)).-....(((....)--))))))).(((((((...)))))))..)).(((.-.....)))........((((.....)))) ( -46.00) >pss.1289 3634006 110 - 6093698 --GAUGGCAUCAA-GGCCCGCGACGCCGGCAGCGGG-CCC-CAACGGCAGAAG-AUUCUGCCCGACCUCAACACGGGGUCUCAGGGAUA-UUACAGUCAACAGACGAGGGCAUUACC-- --((((.(.((..-(((((((..(....)..)))))-))(-(...((((((..-..)))))).((((((.....))))))...))....-.....(((....))))).).))))...-- ( -44.20) >pel.980 4056794 111 - 5888780 --GGCGGCACCUUCAGCCCGCCACGCCGGCAGCGGG-CGCACACCGGCAGAACGAUUCUGCCCGGCCUCGGA--GAGGCCGACGGGACA-UUACAGUCAACAAGUGAGGGCAACACC-- --.(..((.(((...((((((..(....)..)))))-).(((.((((((((.....))))))(((((((...--)))))))..))(((.-.....))).....))))))))..)...-- ( -48.30) >ppk.1205 3793131 111 - 6181863 --GGCGGCACUUUCAGCUCGCCACGCCGGCAGCGAG-CGCAAACCGGCAGAACGACUCUGCCCGGCCUCGGA--GAGGCCGACGGGACA-UUACAGUCAACAAGUGAGGGCAACACC-- --((((((.......)).))))..(((((..((...-.))...)))))..........(((((((((((...--))))))(((.(....-...).))).........))))).....-- ( -44.50) >pfo.1021 4449300 112 - 6438405 --GUCAACACCUU-GGCUCGCCACGCCGGCAGCGAGCCGC-CAACGGCAGAGUUUUUCUGCCAGACCUCAGUU-GAGGUCGACGCGACAAAAACAGUCAACAAGUGAGGGAAACACC-- --.......((((-(((((((..(....)..)))))))((-....(((((((...))))))).((((((....-))))))...))(((.......))).......))))........-- ( -41.90) >pst.1402 3839454 110 - 6397126 --UAUGGCAUCAA-GGCCCGCCACGCCGGCAGCGGG-CCC-CAACGGCAGAAG-ACUCUGCCCGACCCCACCACGGGGUCUCAGGGAUA-UUACAGUCAACAGACGAGGGCAUUACC-- --...........-.(((((((.(((.....)))))-)((-(...((((((..-..)))))).((((((.....))))))...)))...-.....(((....)))..))))......-- ( -46.00) >consensus __GGCGGCACCUU_GGCCCGCCACGCCGGCAGCGAG_CCC_CAACGGCAGAAG_ACUCUGCCCGACCUCAGC__GAGGCCGACGGGACA_UUACAGUCAACAAGUGAGGGCAACACC__ .....(((........(((((..(....)..))))).(((.....((((((.....)))))).((((((.....))))))...))).........))).........((......)).. (-33.28 = -31.07 + -2.22)
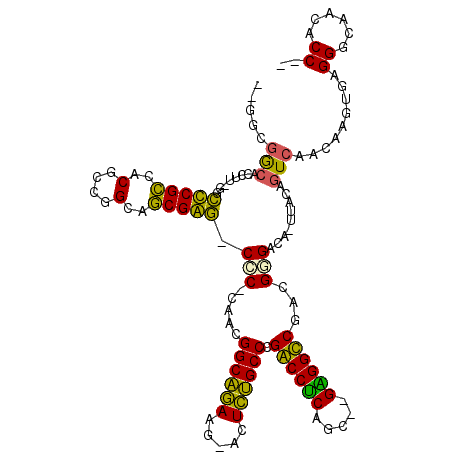
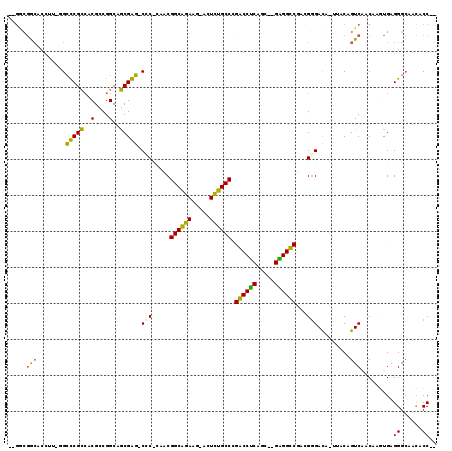
| Location | 3,414,293 – 3,414,400 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.92 |
| Mean single sequence MFE | -49.25 |
| Consensus MFE | -34.25 |
| Energy contribution | -32.53 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1825 3414293 107 - 6264404 AGGGUGUUGCCCUCACUUGUUGACUGUUA-UGUCCCGUUGGCCCCCAGGGGAGGCCUGGCGAAG--CUUUCGCCGAAA-GGG-GUCGUUGCCGGCGUGACGACCC-------UCGCCCG .(((((......((.......(((.....-.))).....((((((....)).)))).(((((..--...)))))))..-(((-(((((..(....)..)))))))-------)))))). ( -46.50) >pss.1289 3634006 110 + 6093698 --GGUAAUGCCCUCGUCUGUUGACUGUAA-UAUCCCUGAGACCCCGUGUUGAGGUCGGGCAGAAU-CUUCUGCCGUUG-GGG-CCCGCUGCCGGCGUCGCGGGCC-UUGAUGCCAUC-- --(((..(((((..(((....))).....-.........((((.(.....).)))))))))..))-)...((.(((..-(((-(((((.((....)).)))))))-)..))).))..-- ( -44.60) >pel.980 4056794 111 + 5888780 --GGUGUUGCCCUCACUUGUUGACUGUAA-UGUCCCGUCGGCCUC--UCCGAGGCCGGGCAGAAUCGUUCUGCCGGUGUGCG-CCCGCUGCCGGCGUGGCGGGCUGAAGGUGCCGCC-- --((((..(((.(((......(((.....-.)))((..(((((((--...)))))))((((((.....)))))))).....(-(((((..(....)..))))))))).)))..))))-- ( -54.00) >ppk.1205 3793131 111 + 6181863 --GGUGUUGCCCUCACUUGUUGACUGUAA-UGUCCCGUCGGCCUC--UCCGAGGCCGGGCAGAGUCGUUCUGCCGGUUUGCG-CUCGCUGCCGGCGUGGCGAGCUGAAAGUGCCGCC-- --((((..((..(((...((.((((....-........(((((((--...)))))))(((((((...))))))))))).))(-(((((..(....)..)))))))))..))..))))-- ( -48.40) >pfo.1021 4449300 112 + 6438405 --GGUGUUUCCCUCACUUGUUGACUGUUUUUGUCGCGUCGACCUC-AACUGAGGUCUGGCAGAAAAACUCUGCCGUUG-GCGGCUCGCUGCCGGCGUGGCGAGCC-AAGGUGUUGAC-- --........(..(((((((..((...............((((((-....)))))).((((((.....))))))))..-))(((((((..(....)..)))))))-.)))))..)..-- ( -49.00) >pst.1402 3839454 110 + 6397126 --GGUAAUGCCCUCGUCUGUUGACUGUAA-UAUCCCUGAGACCCCGUGGUGGGGUCGGGCAGAGU-CUUCUGCCGUUG-GGG-CCCGCUGCCGGCGUGGCGGGCC-UUGAUGCCAUA-- --(((..(((((..(((....))).....-.........((((((.....)))))))))))....-........((..-(((-(((((..(....)..)))))))-)..)))))...-- ( -53.00) >consensus __GGUGUUGCCCUCACUUGUUGACUGUAA_UGUCCCGUCGACCCC__GCUGAGGCCGGGCAGAAU_CUUCUGCCGUUG_GCG_CCCGCUGCCGGCGUGGCGAGCC_AAGGUGCCGCC__ ..((.....))..((((................(((...((((((.....)))))).((((((.....)))))).....))).((((((((....)))))))).....))))....... (-34.25 = -32.53 + -1.72)
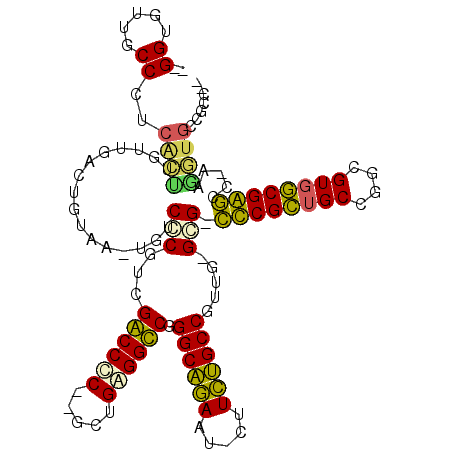
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:56 2007