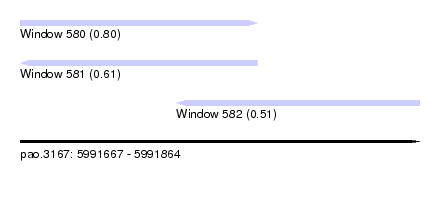
| Sequence ID | pao.3167 |
|---|---|
| Location | 5,991,667 – 5,991,864 |
| Length | 197 |
| Max. P | 0.798239 |

| Location | 5,991,667 – 5,991,784 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -61.60 |
| Consensus MFE | -38.33 |
| Energy contribution | -40.83 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3167 5991667 117 + 6264404 CGCCGAAGCCUACAAGGUCGGCCAGCGCUGGUUGCUGUACAGCGCCGCGCCGGUGCG---CGGCGCCGACCAGAGCCUGCAAGGCACGCUGCUGCUGGCGGUCGACCUGCAACGGCUGCU .((((..((.....((((((((((((((((((((.......(((((((((....)))---))))))))))))).(((.....))).))))((.....)))))))))))))..)))).... ( -65.21) >pss.139 5859568 120 - 6093698 CCCGGAGGCGCGCAAGGUCGGCGAGCGCUGGUUGAUCUACAGCGCUGCGCCGGUGCGUGCCGCUGCCAGUCAGCCGGUGGUGGGUACGCUGCUGCUGGUCUUUGACCUGCAGCGCUUGCU ......((((((((...(((((((((((((.........))))))).))))))))))))))(((((..(((((((((..(..(.....)..)..)))))...))))..)))))....... ( -67.50) >psp.123 5683355 120 - 5928787 CCCGGAGGCUCGCAAGGUGGGCGAGCGCUGGUUGAUCUACAGUGCUGCGCCGGUACGUGCCGCGCCAACUCAGCCGGUGGUGGGCACAUUGCUGCUGGCCUUUGACCUGCAGCGCUUGCU ...........(((((...(((((((((((.........))))))).)))).....(((((.(((((.((.....))))))))))))...(((((.((.......)).))))).))))). ( -53.10) >pst.49 6291270 120 - 6397126 CCCGGAGGCUCGCAAGGUCGGUGAGCGCUGGCUGAUCUACAGCGCAGCGCCGGUACGUGCGGGCGCAGAUCAGCCGGUUCUGGGUACAUUGCUGCUGGCCUUCGACAUGCAGCGCUUGCU ((((((.((((((.......))))))((((((((((((...((((.((((......))))..))))))))))))))))))))))......(((((((........)).)))))....... ( -60.60) >consensus CCCGGAGGCUCGCAAGGUCGGCGAGCGCUGGUUGAUCUACAGCGCUGCGCCGGUACGUGCCGCCGCAAAUCAGCCGGUGCUGGGCACACUGCUGCUGGCCUUCGACCUGCAGCGCUUGCU ..((((((((.(((.(((((((((((((((.........))))))).)))))((((.((((((((.........)))))))).)))))))..))).))))))))....(((.....))). (-38.33 = -40.83 + 2.50)

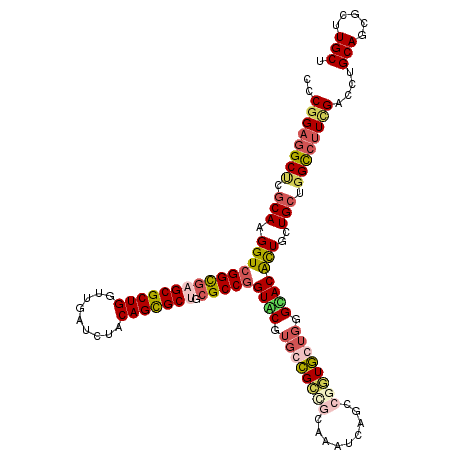
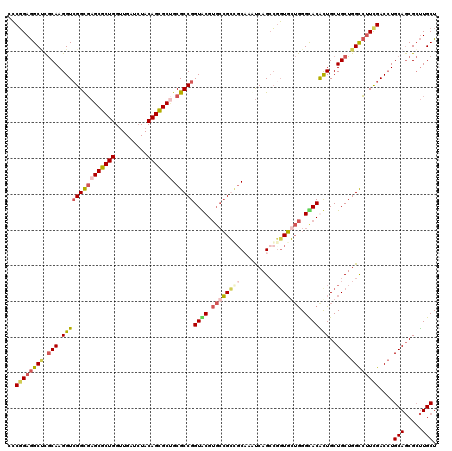
| Location | 5,991,667 – 5,991,784 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -55.62 |
| Consensus MFE | -33.06 |
| Energy contribution | -35.12 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3167 5991667 117 - 6264404 AGCAGCCGUUGCAGGUCGACCGCCAGCAGCAGCGUGCCUUGCAGGCUCUGGUCGGCGCCG---CGCACCGGCGCGGCGCUGUACAGCAACCAGCGCUGGCCGACCUUGUAGGCUUCGGCG ....((((((((((((((...((((((.(((((.(((...))).)))(((..((((((((---(((....)))))))))))..)))......))))))))))))).)))))....)))). ( -66.10) >pss.139 5859568 120 + 6093698 AGCAAGCGCUGCAGGUCAAAGACCAGCAGCAGCGUACCCACCACCGGCUGACUGGCAGCGGCACGCACCGGCGCAGCGCUGUAGAUCAACCAGCGCUCGCCGACCUUGCGCGCCUCCGGG .....(((((((.((((...)))).))))).))..........(((((((.....))))(((.((((.(((((.(((((((.........))))))))))))....)))).)))..))). ( -58.20) >psp.123 5683355 120 + 5928787 AGCAAGCGCUGCAGGUCAAAGGCCAGCAGCAAUGUGCCCACCACCGGCUGAGUUGGCGCGGCACGUACCGGCGCAGCACUGUAGAUCAACCAGCGCUCGCCCACCUUGCGAGCCUCCGGG .(((((.(((((.(((.....))).))))).((((((((.(((.(......).))).).)))))))...((((.(((.(((.........))).)))))))...)))))........... ( -48.70) >pst.49 6291270 120 + 6397126 AGCAAGCGCUGCAUGUCGAAGGCCAGCAGCAAUGUACCCAGAACCGGCUGAUCUGCGCCCGCACGUACCGGCGCUGCGCUGUAGAUCAGCCAGCGCUCACCGACCUUGCGAGCCUCCGGG .(((...(((((.((.(....).)))))))..))).(((.((...((((((((((((..(((((((....))).)))).))))))))))))...((((.(.......).)))).)).))) ( -49.50) >consensus AGCAAGCGCUGCAGGUCAAAGGCCAGCAGCAACGUACCCACCACCGGCUGAUCGGCGCCGGCACGCACCGGCGCAGCGCUGUAGAUCAACCAGCGCUCGCCGACCUUGCGAGCCUCCGGG .......(((((.(((.....))).))))).............(((((((.....))))(((.((((.(((((.(((((((.........))))))))))))....)))).)))..))). (-33.06 = -35.12 + 2.06)

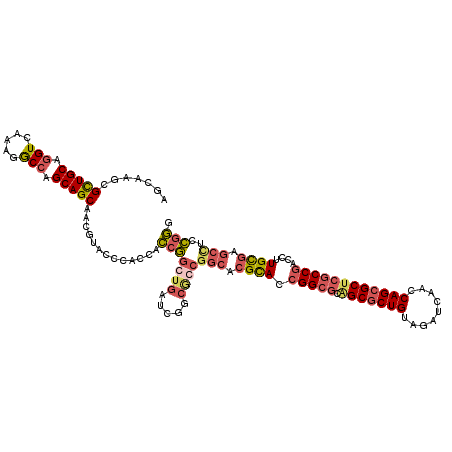
| Location | 5,991,744 – 5,991,864 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.81 |
| Mean single sequence MFE | -53.17 |
| Consensus MFE | -38.09 |
| Energy contribution | -38.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3167 5991744 120 - 6264404 GCACCUGGGCCGGUGCGCCGGAGAACUGCUGGACCAGUUGUACCUGGCCGGCAUCGCCCGGCAAGGGCGGCAGGGCGCGCAGCAGCCGUUGCAGGUCGACCGCCAGCAGCAGCGUGCCUU ......((((((.((((((((.((..((((((.((((......))))))))))))..)))))...(((((..((.(..(((((....))))).).))..)))))....))).)).)))). ( -62.60) >pss.139 5859648 120 + 6093698 ACACCUGAGCAGGUCCGCUGCCGAAUUGCUGGCUGAGCACCACUUGCCCGGCGUCGGCAGGCAGCGCCGGCAAGGCAUUGAGCAAGCGCUGCAGGUCAAAGACCAGCAGCAGCGUACCCA ...........(((.((((((.....(((((((((.(((.....))).))))((((((..(((((((..((..........))..)))))))..)))...)))))))))))))).))).. ( -51.60) >psp.123 5683435 120 + 5928787 AAACCUGUGCAGGUCCGUUGCCAAACUGCUGGCUGAGCACCACCUGCCCGGCCUCGGCAGGCAGUGCCGGCAAGGCAUUGAGCAAGCGCUGCAGGUCAAAGGCCAGCAGCAAUGUGCCCA ...........((..((((((.....((((((((....(((.((((((.......))))))(((((((.....))))))).(((.....))).)))....))))))))))))))..)).. ( -52.40) >pst.49 6291350 120 + 6397126 AAACCUGCGCAGGUCCGCUGCCGAACUGCUGACUGAGGACGAUUUGCCCAGCCUCGGCCGGCAGCACGGGCAAGGCAUCGAGCAAGCGCUGCAUGUCGAAGGCCAGCAGCAAUGUACCCA ..((((....))))..(((((((..((((((.((((((.(..........))))))).))))))..))(((..(((((..(((....)))..)))))....))).))))).......... ( -46.10) >consensus AAACCUGAGCAGGUCCGCUGCCGAACUGCUGGCUGAGCACCACCUGCCCGGCCUCGGCAGGCAGCGCCGGCAAGGCAUUGAGCAAGCGCUGCAGGUCAAAGGCCAGCAGCAACGUACCCA ...........(((.(((((.....(((((((((....(((..(((((.......)))))(((((((..((..........))..))))))).)))....)))))))))))))).))).. (-38.09 = -38.15 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:47 2007